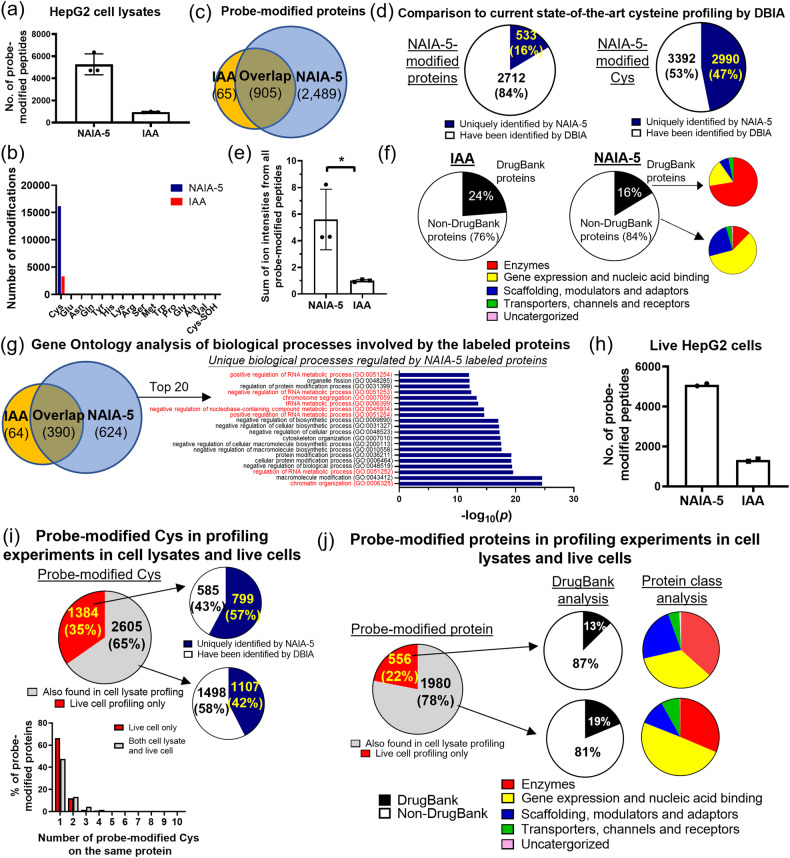
Fig. 7. LC–MS/MS-based chemoproteomics experiments to investigate Cys profiling in HepG2 cell lysates and in live HepG2 cells by NAIA-5 and IAA.
a Number of probe-modified peptides in HepG2 cell lysates identified in the MS experiment outlined in Supplementary Fig. 15. Quantified data were shown in average ± SD from n = 3 different replicates/group. b Modifications on different amino acids by NAIA-5 and IAA, respectively, as identified from the MS experiment. Cys-SOH: sulfenic acid form of Cys. c Number of probe-modified proteins identified in the triplicate experiment. d Comparison of the proteins and cysteines profiled by NAIA-5 with those profiled by an IAA-derived probe, DBIA, in the current state-of-the-art paper for cysteine profiling56. The full list of profiled proteins and cysteines by NAIA-5 can be found in Supplementary Data 2. e Relative intensity of the sum of signals from probe-modified peptides by NAIA-5 or IAA. Quantified data were shown on average ± SD from n = 3 different replicates/groups. Statistical analyses were performed with unpaired two-tailed Student’s t-tests. *P = 0.023. f Analysis of probe-modified proteins by NAIA-5 and IAA, respectively, using the DrugBank database, and the functions of these proteins by Gene Ontology database. g Gene Ontology analysis of the biological processes involving the probe-modified proteins, with the top 20 unique processes regulated by NAIA-5-modified proteins shown in the bar chart. P values were determined by Fisher’s exact test. Those highlighted in red are associated with gene expression and regulation. h Number of probe-modified peptides identified by incubation of live HepG2 cells with NAIA-5 or IAA. Quantified data were the average values from n = 2 different replicates/groups. i Comparison of probe-modified cysteines and j probe-modified proteins identified by NAIA-5 in cell lysates with those in live cell experiment.