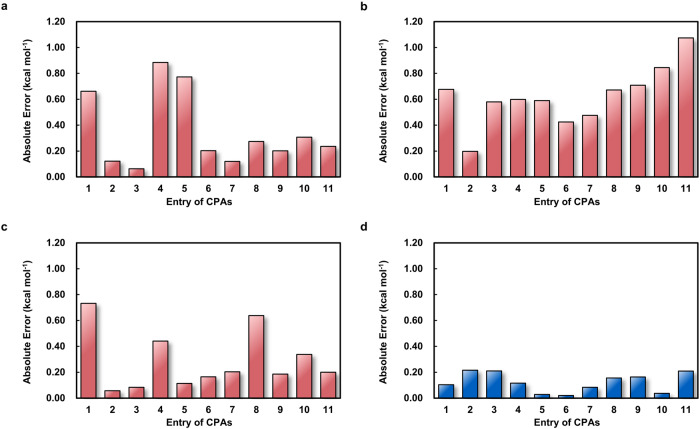
Fig. 7. Errors of predictions for the experimentally tested CPAs by SEMG-MIGNN model (steric- and electronics-embedded molecular graph with molecular interaction graph neural network) and other strategies.
SEMG results are shown in blue. Other strategies’ results are shown in red.a Errors of predictions of the ACSFs-GB model (atom-centered symmetry functions with gradient boosting). b Errors of predictions of the DRFP-XGB model (differential reaction fingerprint with XGBoost). c Errors of predictions of the MFF-RF model (multiple fingerprint feature with random forest). d Errors of predictions of the SEMG-MIGNN model (steric- and electronics-embedded molecular graph with molecular interaction graph neural network). These comparisons demonstrate the advantageous predictions by the SEMG-MIGNN model (steric- and electronics-embedded molecular graph with molecular interaction graph neural network), whose predictions are all within a reasonable error, while the other models encounter pitfall cases where the error of prediction can be larger than 0.5 kcal mol−1. Source data are provided as a Data_for_Fig_7.csv.