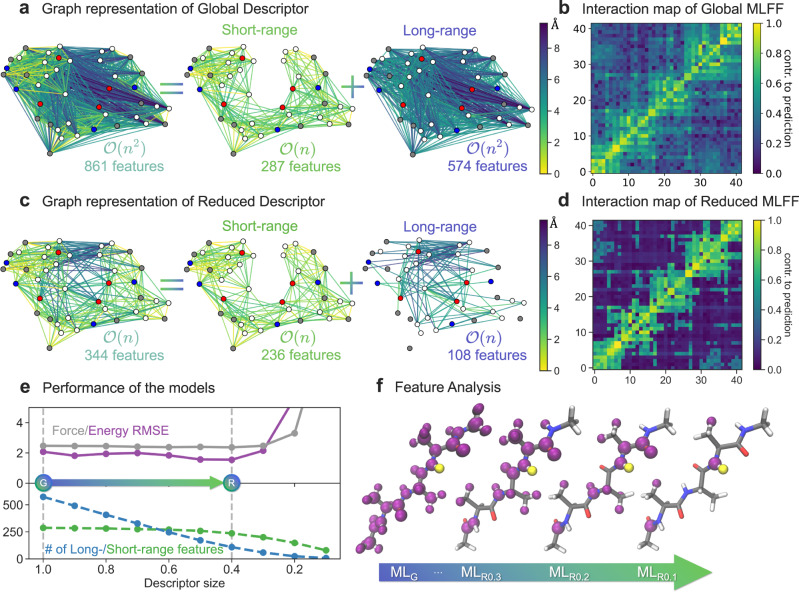
Fig. 1. Overview of the descriptor reduction scheme.
a, c Graph representation of global and reduced descriptors for Ac-Ala3-NHMe, and its decomposition into short- and long-range features with corresponding scaling with the number of atoms, n. The color of nodes indicates atom type: H - white, C - gray, N - blue, O - red. The color of the edges indicates the average distance between atoms. b, d Interaction map of global and reduced Machine Learning Force Fields (MLFFs). Each square in the heatmaps represents a given pair of atoms in the molecule (atom indices start from 0). The colorbar is in log scale normalized to the 0-1 range and goes from dark magenta (small) to yellow (large) for the average contributions to the force prediction. e Performance of the global and reduced models: energy (in kcal mol−1) and force (in kcal (mol Å)−1) root mean square errors (RMSEs) as a function of the size of the descriptor (upper panel). The RMSE values were calculated on a test set of ~80k points, distinct from the training (1k) and validation sets (1k). Decomposition of the descriptor into short- and long-range features (lower panel). The arrow between two vertical dashed lines highlights the degree of reduction between global (G) and optimally reduced (R) models. Descriptor sizes in x axis go from 1 to 0, where 1 corresponds to a default global descriptor and 0 to an empty descriptor. f Feature analysis in the global (denoted as MLG) and reduced models (denoted as MLRX, where X indicates the descriptor size). Hydrogen atom highlighted in yellow keeps interactions with atoms highlighted in purple. The arrow indicates transition of the employed descriptor from default global to substantially reduced ones.