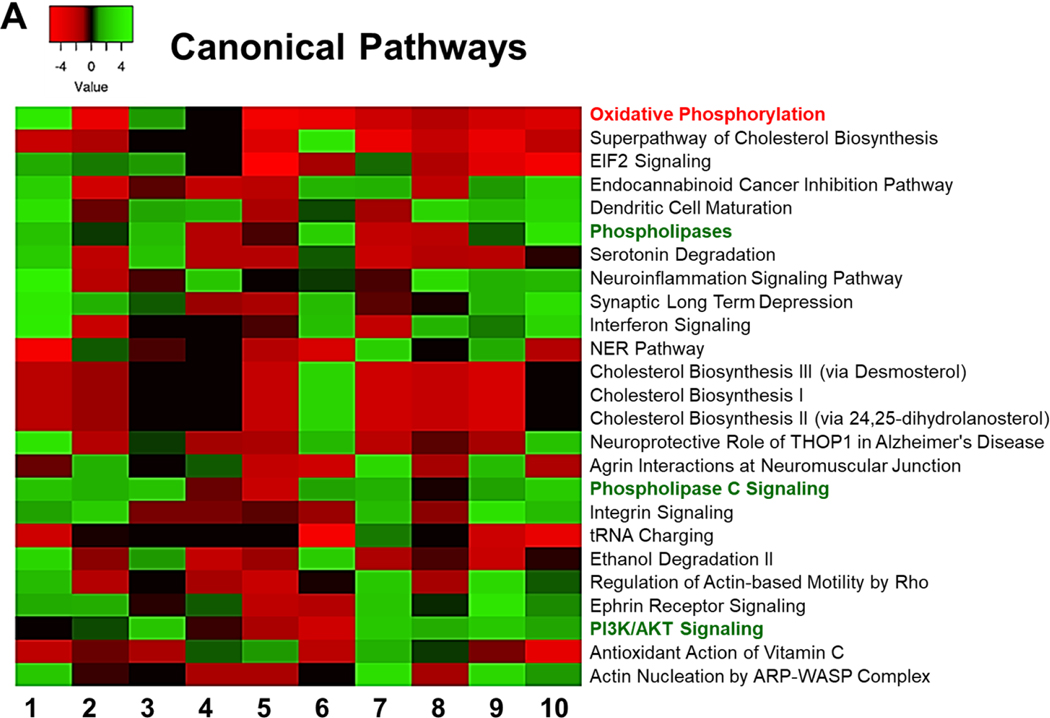
Figure 2: Comparison Analysis.
IPA was used to identify the pathways that are affected by differentially expressed genes in each dataset. A comparison analysis was run to compare the obtained differential gene expression data from each dataset. Affected canonical pathways are sorted according to their average Z scores.