Figure 3.
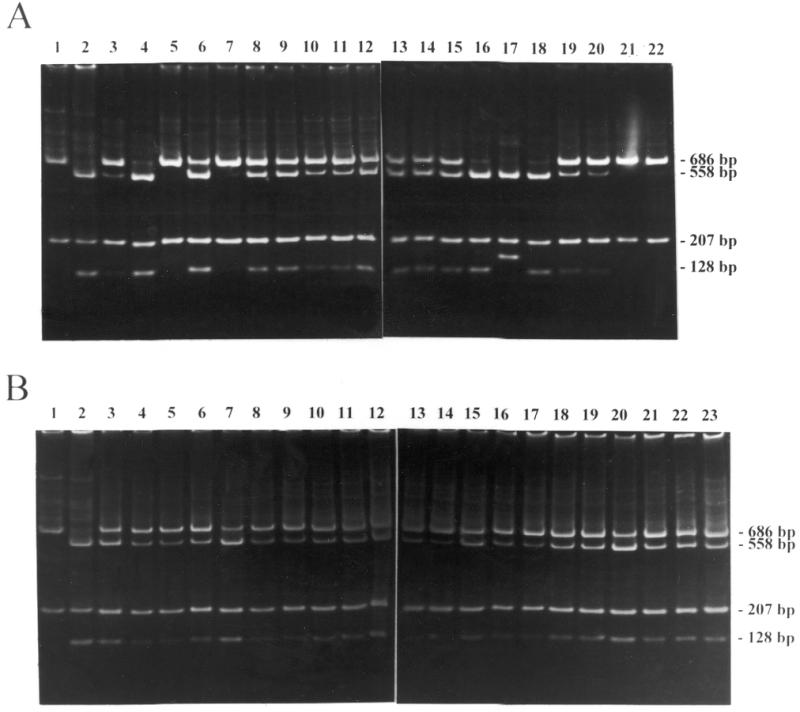
PCR and SmaI digestion analyses of the PBS within the proviral DNA isolated from HeLa (A) and HCT 116 cells (B) infected with LT5. LT5 carrying a T→C mutation within the PBS were used to infect mismatch repair-competent HeLa cells and mismatch repair-defective HCT 116 cells. Each infected cell formed a colony. The PBS from the cells within this colony was amplified by PCR and the PCR products were digested with SmaI. The amplified fragments containing wild-type PBS carry a SmaI site within the R region so that digestion with SmaI results in two fragments of 686 and 207 bp in length (lane 1). The fragments containing mutant PBS carry two SmaI sites so that digestion with SmaI results in three fragments of 558, 207 and 128 bp in length (lane 2). If a colony has a mixture of the wild-type and mutant PBS, digestion with SmaI results in four fragments of 686, 558, 207 and 128 bp in length. All lanes contain SmaI-digested PCR product of DNA isolated from individual neor colonies infected with LT5. In (A) lane 17 contains three fragments of 558, 207 and 163 bp in length. Sequence analysis indicates that a 34 bp sequence (from position 687 to 720) was duplicated at position 687 with an additional SmaI site (CCCGGG) within the PBS. Therefore, sample 17 is a mutant.
