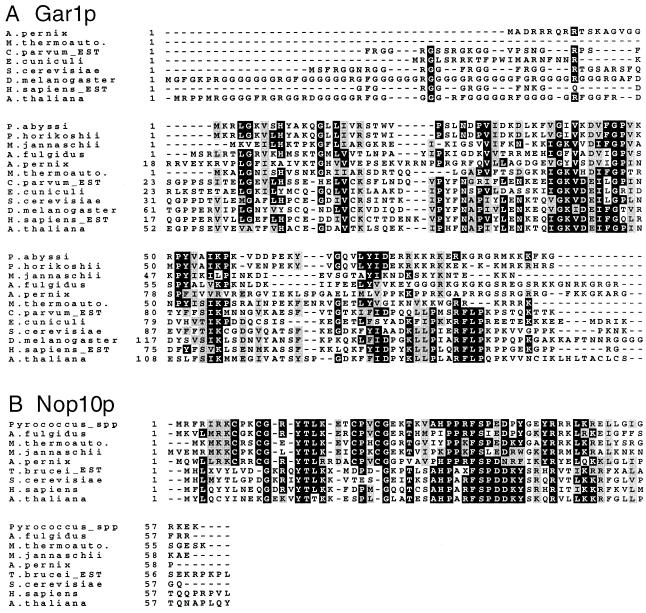
Figure 5.
(A) Alignment of the N-terminal portion of putative Gar1p protein sequences from archaebacteria and some eukaryotes. GenBank accession numbers for these sequences are: P.abyssi, CAB49230; M.jannaschii, P81312; A.pernix, BAA79764; M.thermoautotrophicum, AAB85384; Encephalitozoon cuniculi, CAA07263; S.cerevisiae, P28007; D.melanogaster, S49193; Arabidopsis thaliana, AAF00626. The sequences of P.horikoshii, A.fulgidus, C.parvum and H.sapiens (the latter two being partial sequences from EST data) are from open reading frames in nucleotide sequences AP000007.1, AE001014, AA532317 and AA308727, respectively. (B) Alignment of putative Nop10p sequences from archaebacteria and some eukaryotes. Accession numbers are: P.abyssi, CAB49761; P.horikoshii, translation of an open reading frame in AP000004; A.fulgidus, O29724; M.thermoautotrophicum, O27362; M.jannaschii, P81303; A.thaliana, AAD25649; Trypanosoma brucei, translation from AA681026 (EST data). The sequences of S.cerevisiae and H.sapiens Nop10p are from (31) whereas the Aeropyrum Nop10p sequence is from an open reading frame that begins with TTG in the nucleotide sequence AP000059. Highly conserved residues are shown in white on a black background whereas gray shading indicates conservative substitutions.