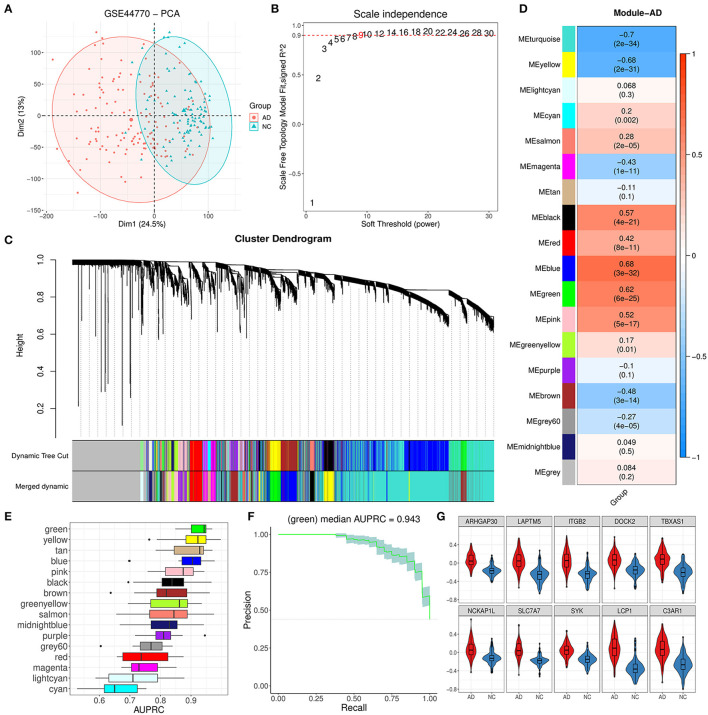
Figure 2.
Module identification by WGCNA and primary exploration. (A) PCA dimension reduction plot for the difference between AD and NC samples in GSE44770. (B) Diagram displaying different soft-thresholding power and corresponding scale-free fit index. (C) Cluster dendrogram of genes with dissimilarity based on the topological overlap before and after merging. (D) Correlation heatmap between module eigengene and AD grouping. Each cell denoted the corresponding correlation and p-value for each module. (E) AUPRC distribution of cross-validation (CV) for 16 valid modules. (F) Precision–recall curve for the green module. The gray area represented the confidence interval of the CV. (G) Expression difference for 10 hub genes of the green module between AD and NC.