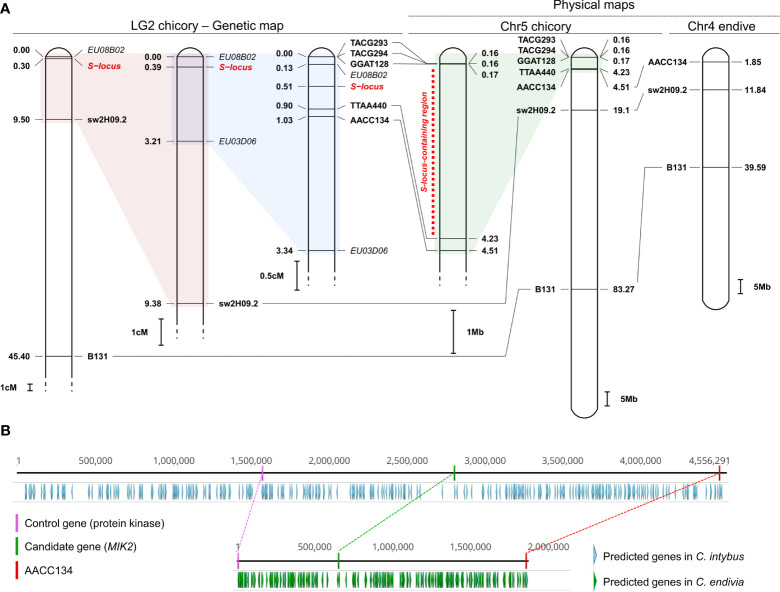
Figure 2.
Narrowing down the chromosomal window containing the SSI locus and identification of a candidate gene. (A) Correspondence between LG2 of Cichorium intybus (Gonthier et al., 2013), chromosome 5 of C intybus and chromosome 4 of C. endivia (Fan et al., 2022). Seven molecular markers (B131 and swH09.2, TACG2942, TACG2932, AACC1342, TTAA4402, GGATT1282, in bold), originally identified by Gonthier et al. because of their association with the S-locus (in red), were mapped against the genome of chicory (JAKNSD000000000), and the SSI-locus was localized in a window of 4 M bases on chromosome 5 (red dotted line). Three out of seven markers were also successfully mapped against the endive genome (JAKOPN000000000), allowing us to identify the correspondence between chromosome 4 and chromosome 5 of chicory. Markers in italics represent other molecular markers identified by Gonthier et al. and associated with the SSI locus whose sequences are not available in NCBI. (B) Comparison between the region of ~4 M bp located on the peripheral arm of chromosome 5 and containing the SSI locus in chicory (upper part) and the corresponding region (~1.8 M) in endive located on the peripheral arm of chromosome 4 (bottom part). Along with all the predicted genes available from Fan et al., (Fan et al., 2022), we highlighted i) the AACC134 marker that delimits the abovementioned chromosomal windows, ii) the newly identified candidate gene (MIK2) in chicory and its putative orthologous in endive, and iii) a control gene (and its putative orthologous in endive) coding for a putative protein kinase, chosen as a control for a comparison between its polymorphism rate and that of the candidate gene.