Figure 1.
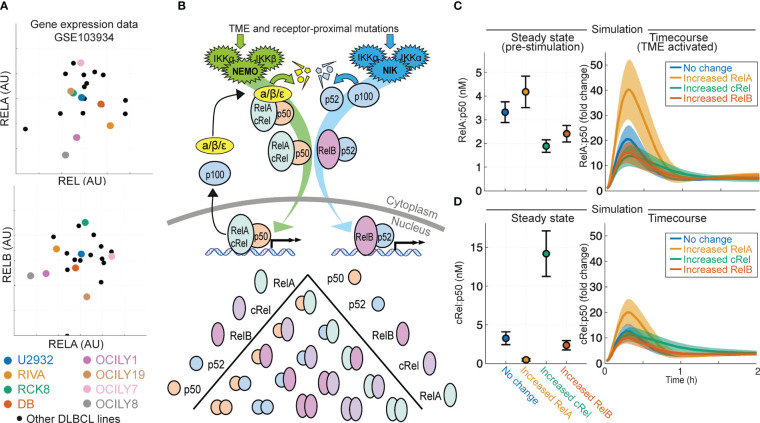
Computational modeling predicts that expression of NF-κB subunit RelA determines response to the tumor microenvironment in DLBCL. (A) Expression of RELA (encoding RelA), REL (encoding cRel) and RELB (encoding RelB) in published gene expression data (GSE103934) for a library of DLBCL cell lines (53). Some well-studied cell lines are highlighted with distinct colors. (B) Schematic of the scope of the computational model used here (54), which includes both canonical and non-canonical NF-κB signaling and dimer formation between 5 NF-κB component proteins. (C, D) Nuclear RelA:p50 (C) and cRel:p50 (D) concentration in computational simulations using model with no change in parameters, and a 10-fold increase in RelA, cRel and RelB expression. Steady state abundances are shown on the left, with time course responses to TME activation shown on the right. Mean and standard deviation of 25 single cell simulations is indicated.