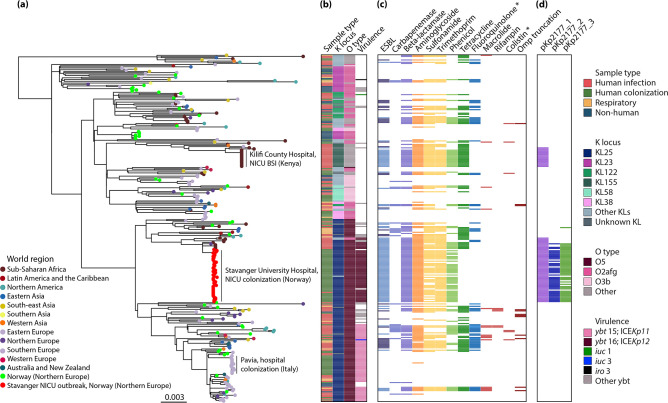
Fig. 3.
Global phylogeny of 300 Klebsiella pneumoniae ST17 genomes. (a) Maximum likelihood tree with tips coloured by region of collection. Additionally, the Stavanger NICU outbreak genomes are coloured red and other genomes from Norway green. The three clonal expansions that were observed in ST17 are labelled. (b) Sample type and loci as indicated in the column names. The most prevalent loci are indicated in the inset legend. (c) Presence (colour) or absence (white) of genes encoding resistance to the listed antimicrobial resistance drug classes (blocks are coloured by drug class). Lighter colour in the ESBL column indicates bla CTX-M-15. (d) Presence (colour) or absence (white) of the Kp2177 plasmids. Several plasmid replicon markers were present across ST17; these are shown in Fig. S4. More details about the metadata and genotypes are available for interactive viewing at https://microreact.org/project/kpst17. *, acquired genes and mutations.