Figure 2. CRISPRme provides analysis of off-target potential of CRISPR-Cas gene editing reflecting population and private genetic diversity.
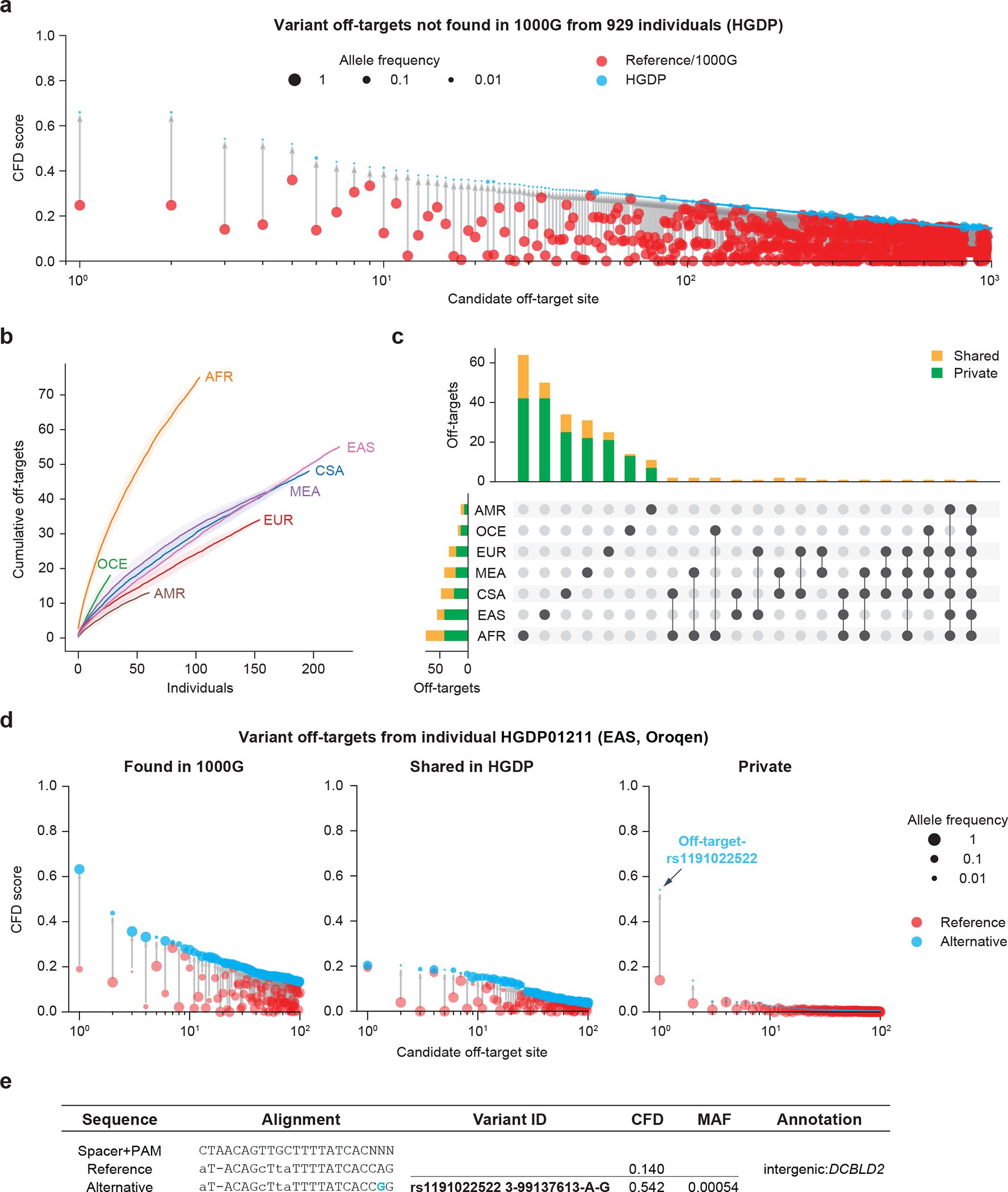
a) CRISPRme analysis was conducted with variants from HGDP comprising whole genome sequencing of 929 individuals from 54 diverse human populations. HGDP variant off-targets with greater CFD scores than the reference genome or 1000G were plotted and sorted by CFD score, with HGDP variant off-targets shown in blue and reference or 1000G variant off-targets shown in red. b) Cumulative distribution plot of HGDP variant off-targets with CFD≥0.2 and increase in CFD of ≥0.1 per super-population. Individual samples from each of the seven super-populations were shuffled 100 times to calculate the mean and 95% confidence interval (shading around lines). c) Intersection analysis of HGDP variant off-targets with CFD≥0.2 and increase in CFD of ≥0.1. Shared variants (orange) were found in 2 or more HGDP samples while private variants (green) were limited to a single sample. d) CRISPRme analysis of a single individual (HGDP01211) showing the top 100 variant off-targets from each of the following three categories: shared with 1000G variant off-targets (left panel), higher CFD score compared to reference genome and 1000G but shared with other HGDP individuals (center panel), and higher CFD score compared to reference genome and 1000G with variant not found in other HGDP individuals (right panel). For the center and right panels, reference refers to CFD score from reference genome or 1000G variants. e) The top predicted private off-target site from HGDP01211 is an allele-specific off-target where the rs1191022522-G minor allele produces a canonical NGG PAM sequence in place of a noncanonical NAG PAM sequence. Spacer shown as DNA sequence for ease of visual alignment.
