Figure 3. Allele-specific off-target editing by a BCL11A enhancer targeting gRNA in clinical trials associated with a common variant in African-ancestry populations.
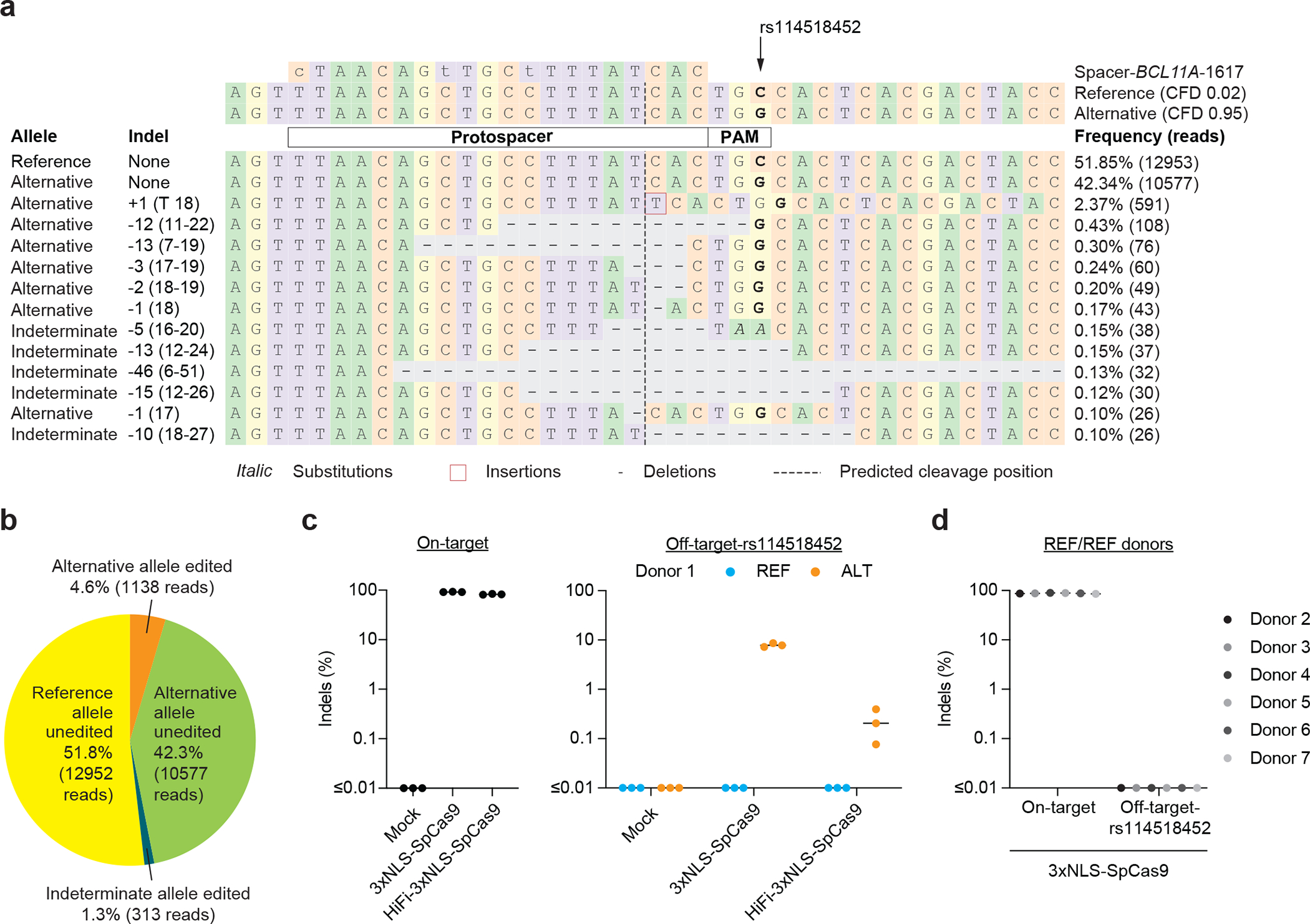
a) Human CD34+ HSPCs from a donor heterozygous for rs114518452-G/C (Donor 1, REF/ALT) were subject to 3xNLS-SpCas9:sg1617 RNP electroporation followed by amplicon sequencing of the off-target site around chr2:210,530,659–210,530,681 (off-target-rs114518452 in 1-start hg38 coordinates). CFD scores for the reference and alternative alleles are indicated and representative aligned reads are shown. Spacer shown as DNA sequence for ease of visual alignment, with mismatches indicated by lowercase and the rs114518452 position shown in bold. b) Reads classified based on allele (indeterminate if the rs114518452 position is deleted) and presence or absence of indels (edits). c) Human CD34+ HSPCs from a donor heterozygous for rs114518452-G/C (Donor 1) were subject to 3xNLS-SpCas9:sg1617 RNP electroporation, HiFi-3xNLS-SpCas9:sg1617 RNP electroporation, or no electroporation (mock) followed by amplicon sequencing of the on-target and off-target-rs114518452 sites. Each dot represents an independent biological replicate (n = 3), lines represent medians. Indel frequency was quantified for reads aligning to either the reference or alternative allele. d) Human CD34+ HSPCs from 6 donors homozygous for rs114518452-G/G (Donors 2–7, REF/REF) were subject to 3xNLS-SpCas9:sg1617 RNP electroporation with 1 biological replicate per donor followed by amplicon sequencing of the on-target and off-target-rs114518452 sites.
