Figure 4. Allele-specific pericentric inversion following BCL11A enhancer editing due to off-target cleavage.
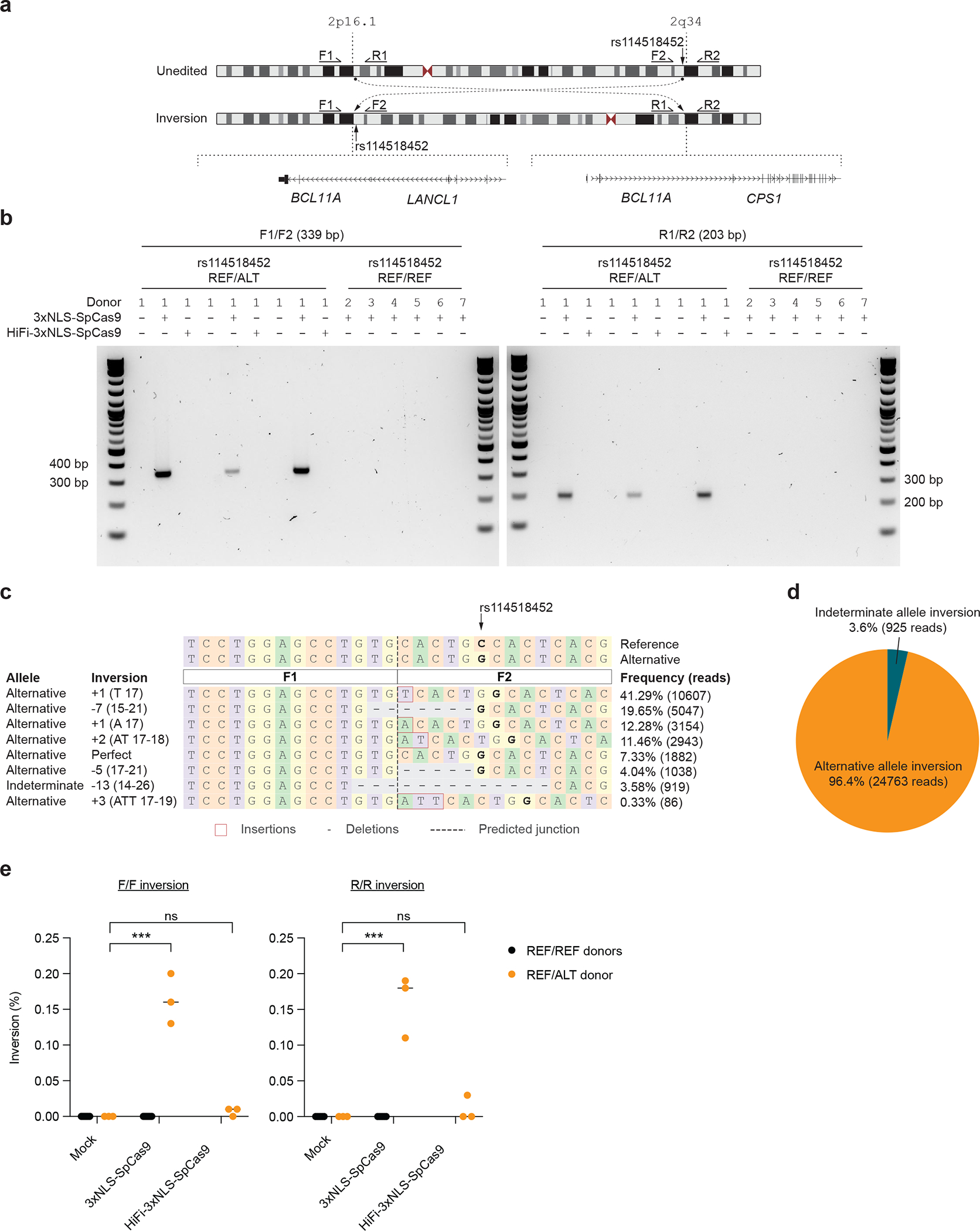
a) Concurrent cleavage of the on-target and off-target-rs114518452 sites could lead to pericentric inversion of chr2 as depicted. PCR primers F1, R1, F2, and R2 were designed to detect potential inversions. b) Human CD34+ HSPCs from a donor heterozygous for rs114518452-G/C (Donor 1) were subject to 3xNLS-SpCas9:sg1617 RNP electroporation, HiFi-3xNLS-SpCas9:sg1617 RNP electroporation, or no electroporation with 3 biological replicates. Human CD34+ HSPCs from 6 donors homozygous for rs114518452-G/G (Donors 2–7, REF/REF) were subject to 3xNLS-SpCas9:sg1617 RNP electroporation with 1 biological replicate per donor. Gel electrophoresis for inversion PCR was performed with F1/F2 and R1/R2 primer pairs on left and right respectively with expected sizes of precise inversion PCR products indicated. c) Reads from amplicon sequencing of the F1/F2 product (expected to include the rs114518452 position) from 3xNLS-SpCas9:sg1617 RNP treatment were aligned to reference and alternative inversion templates. The rs114518452 position is shown in bold. d) Reads classified based on allele (indeterminate if the rs114518452 position deleted). e) Inversion frequency by ddPCR from same samples as in (b) with three replicates from the single REF/ALT donor and one replicate each from the six REF/REF donors. F/F indicates forward and R/R reverse inversion junctions as depicted in (a).
