Figure 5. CRISPRme illustrates prevalent off-target potential due to genetic variation.
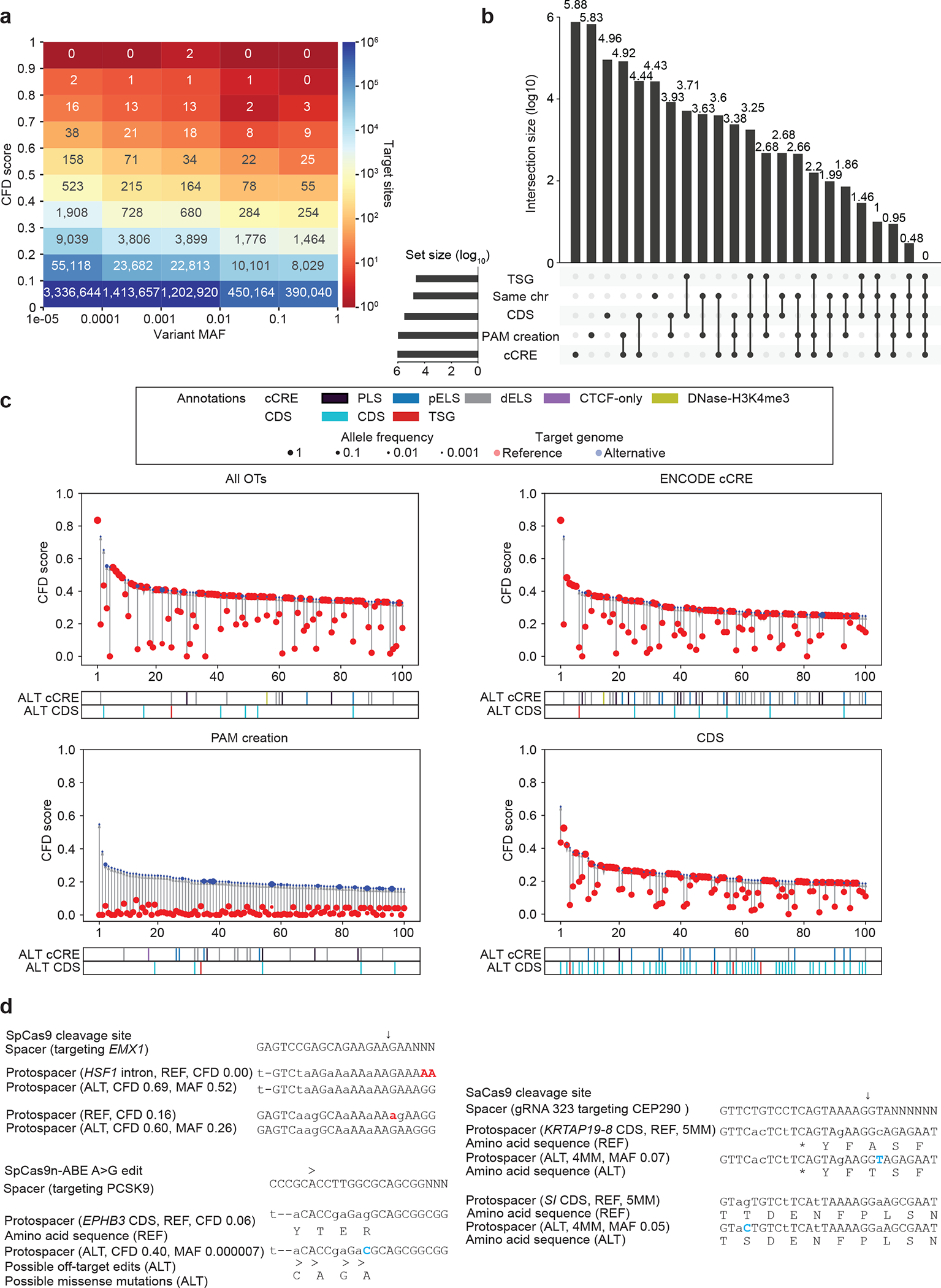
a) Heatmap showing the distribution of alternative allele nominated off-targets for SpCas9 guides by CFD score and MAF. b) UpSet plot showing overlapping annotation categories for candidate off-targets (TSG, tumor suppressor gene; candidate off-targets on the same chromosome as the on-target; CDS regions; cCRE from ENCODE and PAM creation events). c) Top 100 predicted off-target sites ranked by CFD score for the gRNA targeting PCSK9 with no filter, found in cCREs, corresponding to PAM creation events, and in CDS regions) d) Top left: Candidate off-target sites with increased predicted cleavage potential introduced by common (MAF 52% and 26%) indel variants for a SpCas9 gRNA targeting EMX1. Right: Candidate off-target cleavage sites within coding sequences with increased homology to a lead gRNA for SaCas9 targeting of CEP290 to treat congenital blindness in current clinical trials due to common SNPs. Bottom: Potential missense mutations in the EPHB3 tumor suppressor resulting from candidate off-target A-to-G base editing by a preclinical lead gRNA targeting PCSK9 to reduce LDL cholesterol levels. Deletions shown in red, SNPs shown in blue.
