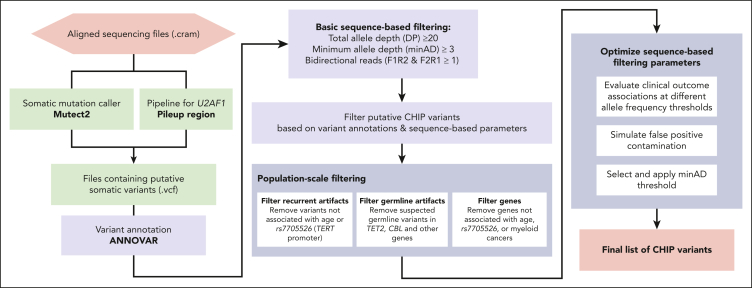
Figure 1.
Schematic of CHIP variant ascertainment workflow. Putative somatic mutations are first identified using a somatic mutation caller and annotated for gene- and protein-level changes. Variants are then filtered based on an initial, liberal set of parameters and filtered based on gene-specific CHIP variant rules. In some genes, all loss-of-function mutations are considered putative CHIP variants, whereas in other genes, only specific missense mutations are included. Leveraging available large-scale sequencing data, we apply 3 filters to identify artifactual genes and variants. We then optimize the sequencing-based filtering parameters, yielding a final CHIP mutation call set.