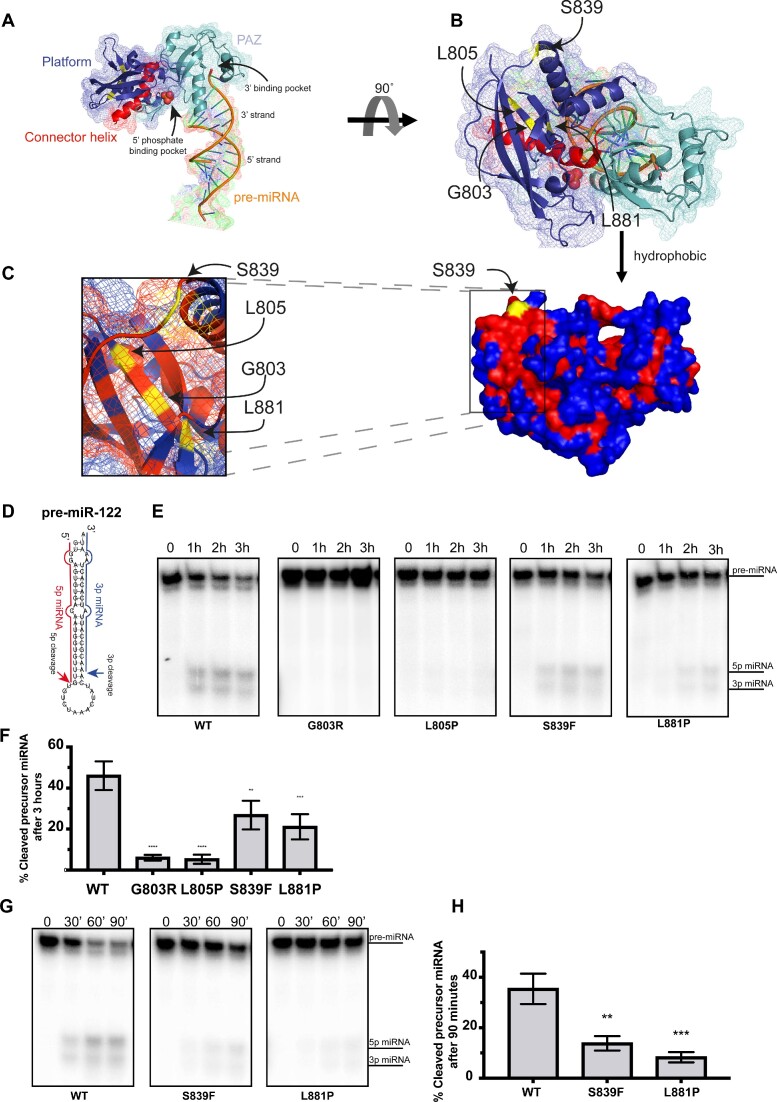
Figure 2.
Positioning and impact of Platform variants on DICER1-mediated pre-miRNA cleavage. (A) Structure of platform (dark blue) and PAZ (cyan) domains with connector helix (red) on crystal structure published by (Tian et al., 2014) (PDB: 4NH3). 5′ phosphate and 3′ binding pocket indicated by arrows. Theoretical pre-miRNA 3′ and 5′ strands are labelled on dsRNA used in crystal structure. (B) Spatial location of platform variants (yellow). (C) Surface model with hydrophobic residues coloured in red and hydrophilic ones in blue. Pathogenic variants are highlighted in yellow. (D) Structure of pre-miR-122 used in in vitro cleavage assay with indicated cleavage events. (E) Auto-radiograph of in vitro cleavage (IVC) assay for DICER1 Platform variants over 1 h time intervals using pre-miR-122. (F) Quantification of the percentage of cleaved product for each IVC reaction after 3 h compared to WT DICER1 (C) (n = 3). (G) Auto-radiograph of in vitro cleavage (IVC) assay for DICER1 Platform mutants over 30 min time intervals. (H) Quantification of the percentage of cleaved product for each IVC reaction compared to WT DICER1 after 90 min (D) (n = 3). Data information: In (D, F), data are presented as mean ± SD. *: 0.05 ≥ P-value > 0.01, **: 0.01 ≥ P-value > 0.001, ***: 0.001 ≥ P-value > 0.0001, ****: 0.0001 ≥ P-value (one-way ANOVA with Dunnett's multiple comparisons test compared to WT DICER1 cleavage).