Figure 2: Technological advances in studying network interactions of helminths, microbes, and host.
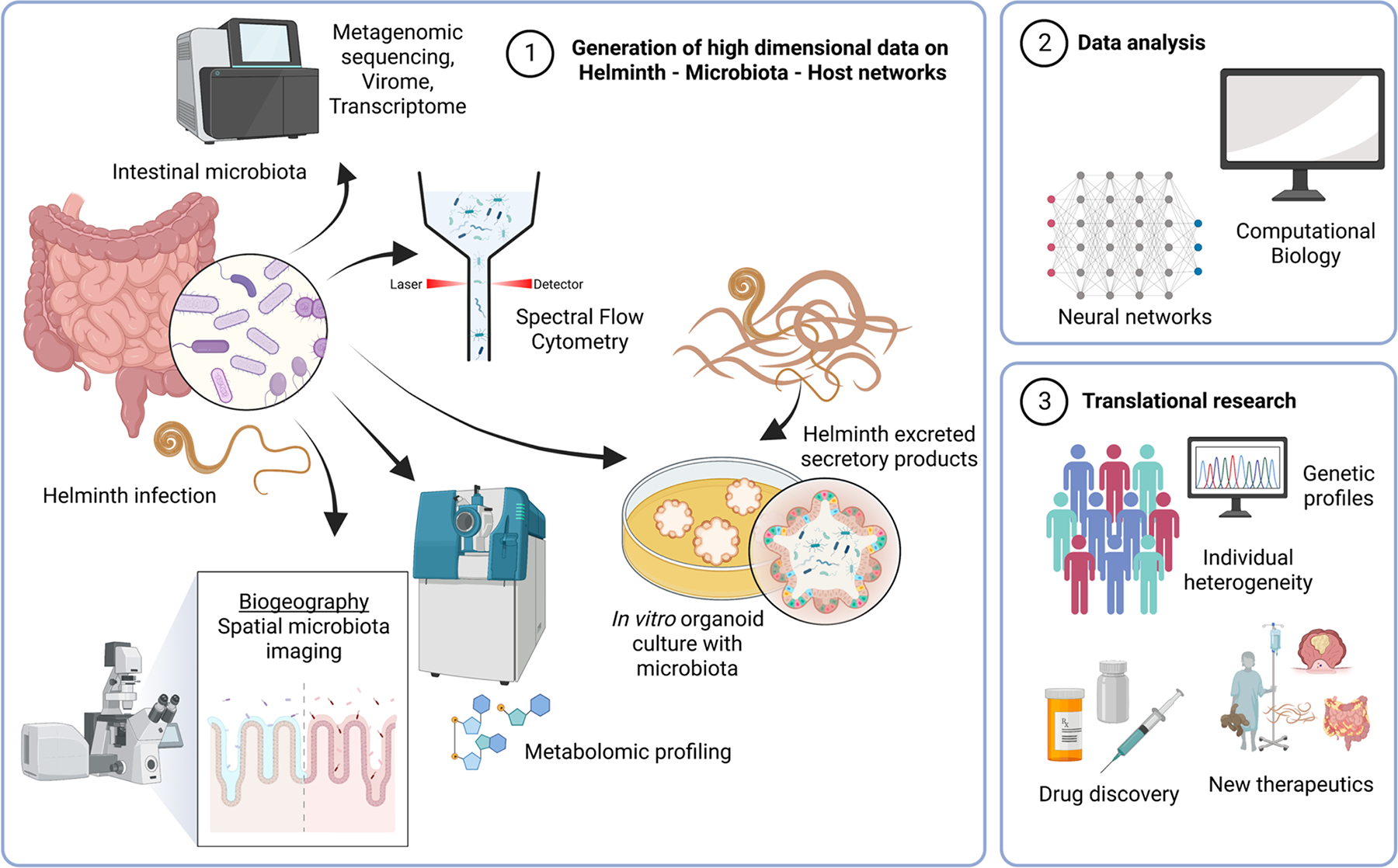
Advances in high dimensional technologies and computational approaches are driving forces in studying the interactions of microbes and helminths with the host immune system. As intestinal helminths and the gut microbiota occupy the same ecological niche, the host intestines have been the major focus of this work. Sequencing technology is a primary driver of many analytical approaches, with newer strategies providing a comprehensive picture of the metagenome, virome and transcriptome of the intestinal microbiota. Other developments include the use of flow cytometry, which can provide absolute count data, as well as sort bacterial populations of interest. Metabolomic profiling provide insights into the biochemical language exchange between host, bacteria and helminths. New imaging and sequencing approaches could enable a better understanding of the biogeography of helminth-bacterial interactions in the intestine. Intestinal biopsies can be used to generate organoid cultures, providing in vitro assay systems to study interactions between helminth secreted molecules and epithelial cells. Bacteria can also be introduced into the organoids in this reductionist system. These high dimensional datasets will then have to undergo extensive data analysis with modern computational biology approaches, including neural network strategies to discern interaction networks. Finally, this information needs to be translated to the clinical setting where there is greater heterogeneity at the host level with many different genetic profiles but has the promise of developing better therapeutic strategies against helminth infections, as well as inflammatory diseases. “Created with BioRender.com”.
