Figure 2.
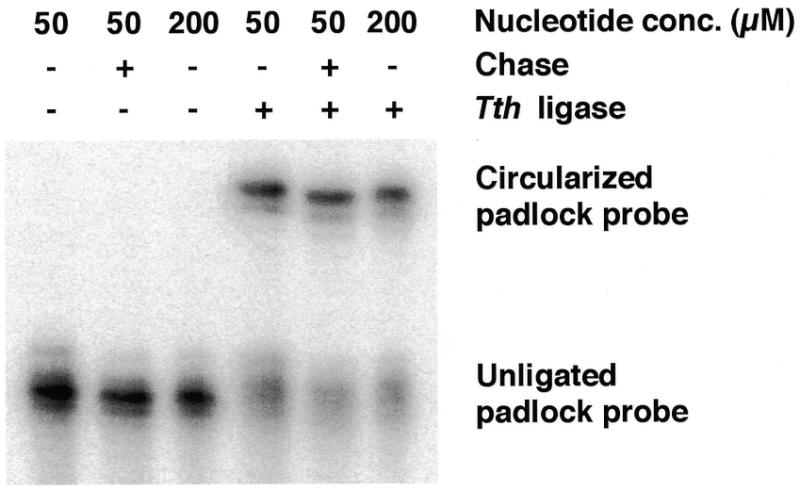
Ligation of PCR-generated padlock probes analyzed by denaturing polyacrylamide gel electrophoresis. Padlock probes were generated using chrAB and chrA as primers and link oligonucleotide as template. DNA was labeled during PCR by addition of 20 µCi [32P]dCTP (NEN, Boston, MA) to the reactions. No extra dNTP was added to the non-chase sample and to a control that was run with 200 µM each dNTP. The padlock probe was circularized using a 2-fold excess (0.5 µM) of the oligonucleotide 5′-AATAGAAATGTTCAACTCCTTTAGCTGGGTACACACATCA-3′ as template in a 10 µl reaction, The reaction was stopped by adding 10 µl of loading buffer containing 10 mM EDTA. The samples were run on a 6% denaturing polyacrylamide gel (Bio-Rad Laboratories, Hercules, CA) and quantified using a PhosphorImager system (Molecular Dynamics, Sunnyvale, CA).
