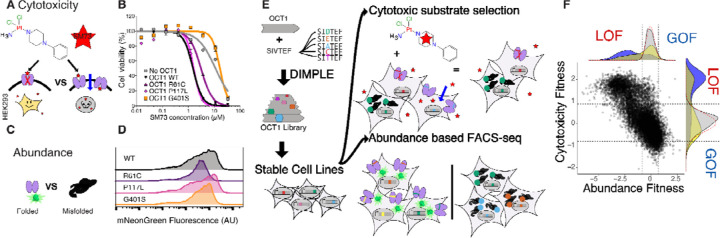
Figure 1. Workflow for multiparametric deep mutational scan of OCT1.
(A) Cytotoxic OCT1 substrates such as SM73 enable negative selection screens for OCT1 function in HEK293 cells. (B) OCT1 variants have different sensitivity to Cisplatin analog SM85, allowing a fitness gradient. (C) A split fluorescent protein-based readout for variant abundance can be used to distinguish folded vs misfolded forms of OCT1 as demonstrated (D) in flow cytometry experiments where loss of transport variants from cytotoxicity screening have diminished fluorescence. (E) We generated an OCT1 deep mutational scanning library with our DIMPLE protocol, produced stable cell lines, and conducted parallel abundance and cytotoxicity selection screens to determine the functional impacts of variants, yielding (F) a multiparametric fitness landscape for 11,213 OCT1 mutants: x-axis, abundance score, and y-axis, cytotoxicity scores, with density plots indicating classes of mutations including synonymous (grey), missense (yellow), and single codon deletion (purple). Cutoffs for loss and gain of function for both phenotypes are indicated by a dotted line, which were determined using a 2 standard deviations from normal distribution fit upon synonymous variant’s distributions.