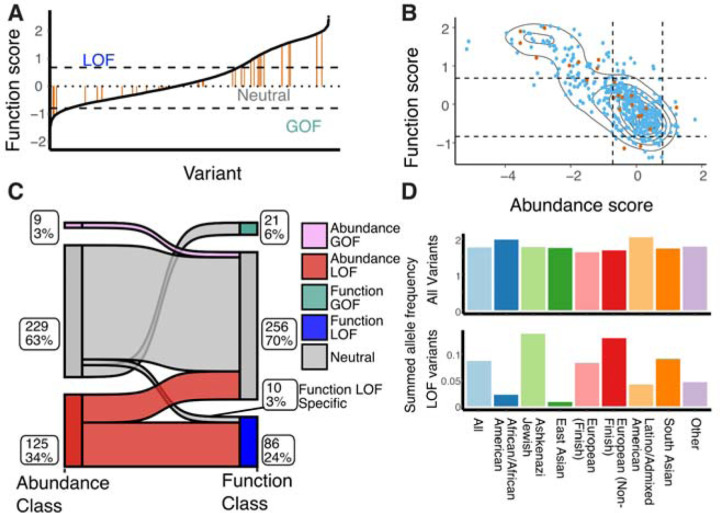
Figure 6. Mechanism of action of OCT1 polymorphisms.
(A) Distribution of previously characterized human polymorphisms from gnomAD (39 variants, orange) among the total set from this work is shown among the rank-ordered set of all 515 gnomAD variants. Cutoffs are shown based on the synonymous mutation distributions as dotted lines. (B) Abundance and function scores plotted for all human variants, showing previously-studied variants (orange) among all previously uncharacterized (blue). Cutoffs are shown based on synonymous mutation distribution as dotted lines. (C) Classification of human variants by their predicted impact based on synonymous-derived cutoffs. The diagram shows how observed functional variation (right) is conditioned on abundance impacts (left). Numbers and percentages of variants within each group are next to the respective subpopulation. Flows between classes suggest the interaction between abundance and function. (D) Summed allele frequencies across populations with differing ancestry within gnomAD reveal the utility of functional annotation for understanding human variation. Across all variants, all groups have similar alternative allele frequencies (top), but looking at only loss of function variants reveals large differences (bottom).