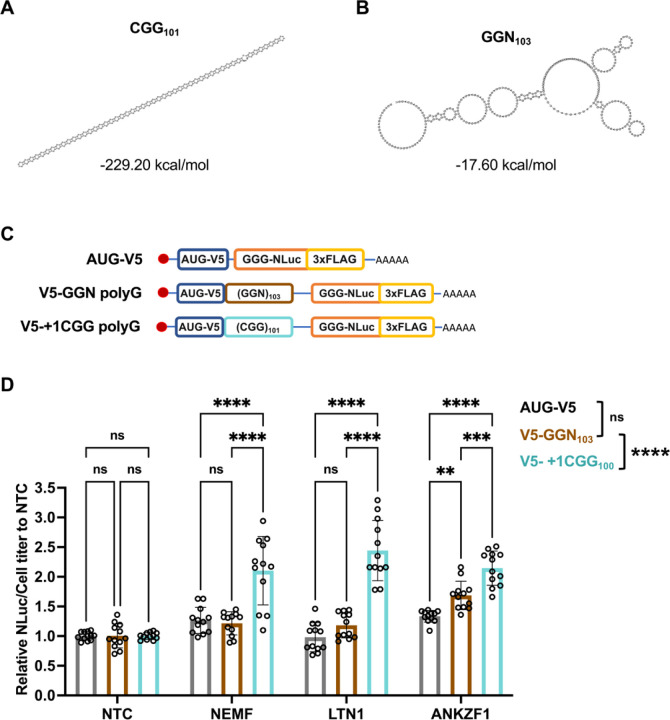
Figure 5. Enhancement of polyG production with NEMF, LTN1, and ANKZF1 depletion requires the CGG repeat RNA structure.
(A-B) Prediction of the optimal RNA secondary structure and calculation of the minimum free energy in CGG101 and GGN103 repeats. Only the repeat region from each repeat was used to predict the RNA secondary structure and calculate the minimum free energy. The results were computed by RNAfold 2.5.1. (C)Schematics of AUG-V5-NLuc-3xFLAG, AUG-V5-+1(CGG)101-NLuc-3xFLAG, and AUG-V5-(GGN)103-NLuc-3xFLAG transcripts. (D) Knockdown of NEMF, LTN1, and ANKZF1 in HEK293 with no-repeat control, polyG from GGN repeats, and CGG repeats RNA transfection. Data represent means with error bars ±SD of n = 12, ns = not significant; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001 by Tukey’s multiple comparisons tests. The statistic result placed in the legend is group comparisons by two-way ANOVA Sidak’s multiple comparisons tests.