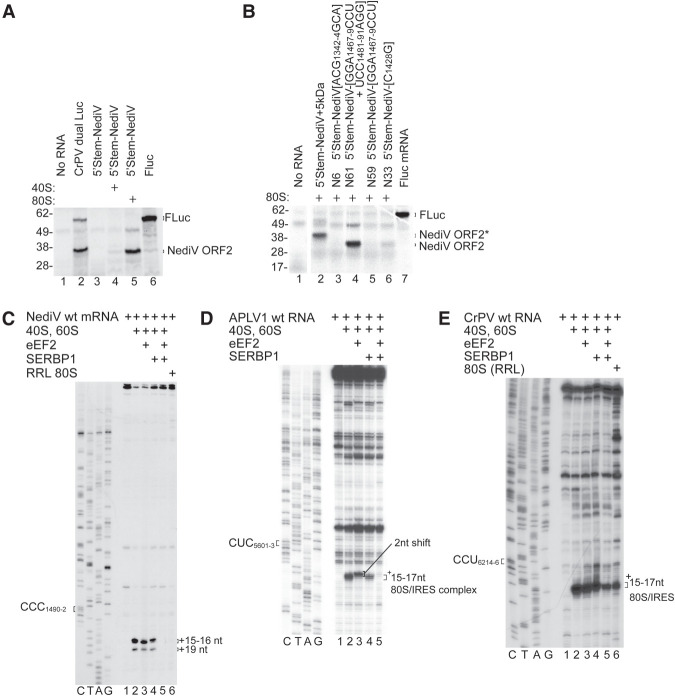
FIGURE 4.
Inhibition of NediV IGR IRES function by SERBP1/eEF2. (A,B) Translation of 5′-stem-NediV IRES mRNAs in RRL with or without preincubation with purified 40S subunits or 80S ribosomes. (A) FLuc and CrPV dual Luc mRNAs are positive controls. The FLuc and NediV ORF2 translation products are indicated on the right and molecular weight markers (kDa) on the left. (B) Translation of 5′-stem-NediV IRES + 5 kDa and mutant versions of 5′-stem-NediV IRES mRNA with substitutions in IRES domain 1 (N6), domain 2 (N33), and domain 3 (N59 and N61), after preincubation with 80S ribosomes. FLuc mRNA was a positive control. The FLuc, NediV ORF2, and NediV-(ORF2 + 5 kDa) translation products are indicated on the right and molecular weight markers (kDa) on the left. (C) Influence of SERBP1 and eEF2 on binding of ribosomes to the NediV IRES, assayed by toeprinting. The positions of +16–+17 nt and +19 nt toeprints are shown on the right. Lanes C, T, A, G depict the NediV sequence. (D,E) Influence of SERBP1 and eEF2 on ribosomal complex formation on (D) APLV1 and (E) CrPV IRESs incubated with SERBP1, eEF2, and ribosomal subunits or native ribosomes, as indicated. The positions of the P-site codon and of ribosomal complexes are indicated on the left and on the right of gel panels, respectively. Lanes C, T, A, and G show (D) APLV1 and (E) CrPV sequences, respectively. (B) Separation of lanes by white lines indicates that they were juxtaposed from the same gel.