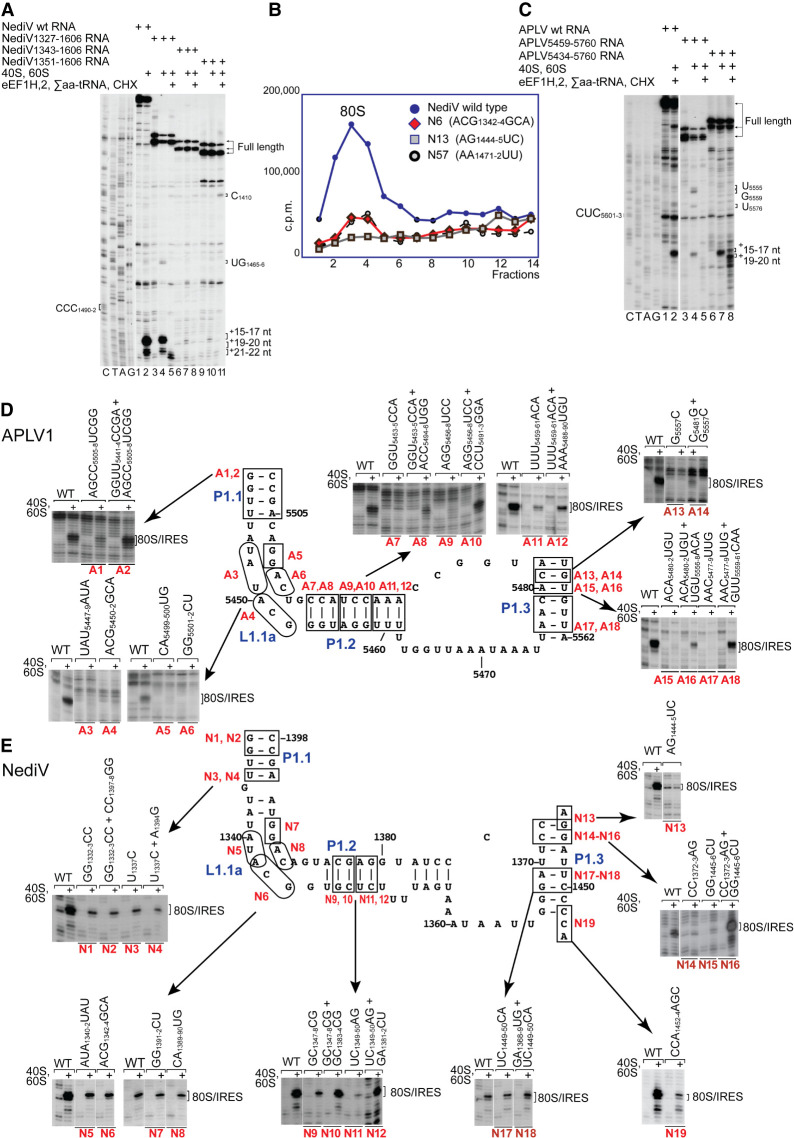
FIGURE 5.
Mutational analysis of domain 1/PKII. (A,C) Toeprinting analysis of the influence of 5′-terminal deletions of (A) the NediV IRES and (B) the APLV1 IRES on their ability to support binding of ribosomes and subsequent elongation upon inclusion of the indicated components. The positions of the codons that occur immediately upstream of ORF2 coding sequences are indicated on the left, and full-length cDNAs and toeprints from ribosomal complexes before and after elongation are indicated on the right. Lanes C, T, A, G depict (A) NediV and (C) APLV1 sequences. (B) Association of 32P-labeled wt and mutant NediV IGR-containing mRNAs with 80S ribosomes, assayed by sucrose density gradient centrifugation. Mutant mRNAs had substitutions in domain 1 (N6, N13) or in domain 3 (N57). Sedimentation was from right to left, and the position of 80S complexes is indicated. Fractions from the upper part of the sucrose gradient have been omitted for greater clarity. (D,E) The influence of disruptive and compensatory substitutions in P1.1, P1.2, and P1.3 helices and the L1.1 loop of (D) APLV1 and (E) NediV IRESs, as indicated on the models of IRES domain 1. IRES activity in ribosomal binding was assayed by toeprinting. The positions of 80S:IRES complexes are indicated on the right of each panel. (A,C–E) Separation of lanes by white lines indicates that they were juxtaposed from the same gel.