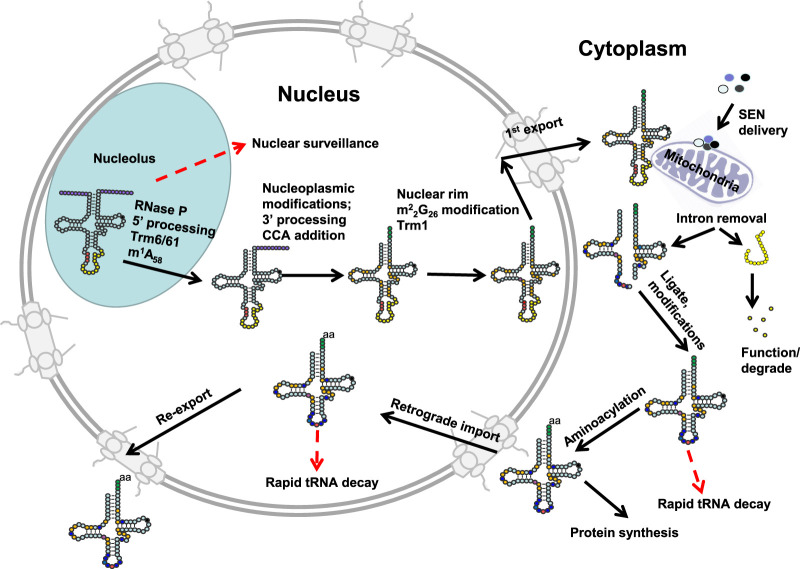
FIGURE 1.
Schematic of tRNA biogenesis, subcellular dynamics, and quality control turnover pathways in S. cerevisiae. tRNAs are transcribed in the nucleolus where the 5′ leader (left purple circles) of the initial transcript is removed by RNase P and likely where m1A58 is modified (black circle) by Trm6/61. About half of the known modifications (examples, orange circles) occur in the nucleoplasm where 3′ CCA nucleotides (green circles) are also added. Dimethylation of G26 (magenta circle) is catalyzed by Trm1, which is located on the inner nuclear membrane, prior to nuclear export of the end-matured, partially processed, intron-containing (yellow circles) pre-tRNAs; end-processed, partially modified tRNAs encoded by genes lacking introns are also exported to the cytoplasm. Introns are removed on the mitochondrial cytoplasmic surface. After/during splicing, additional modifications are added in the cytoplasm (examples, blue circles), and the freed introns are destroyed. Processed/modified cytoplasmic tRNAs return to the nucleoplasm via retrograde tRNA nuclear import and under stress conditions accumulate there; in favorable conditions the tRNAs return to the cytoplasm via reexport where they participate in protein synthesis. There are quality control steps, indicated by red dashed arrows, that destroy tRNAs that have not undergone the canonical (black arrows) steps appropriately. Further details of the cell biology and quality control pathways are provided in the text and Figures 7 and 8.