FIGURE 7.
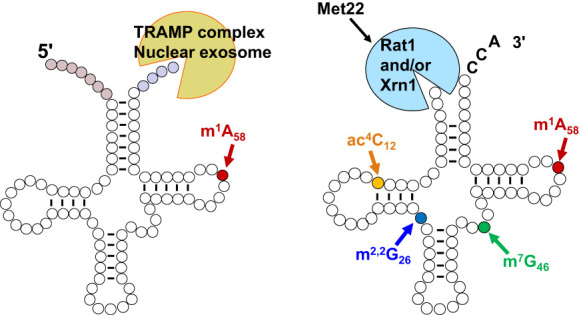
Two different tRNA decay pathways in S. cerevisiae. (Left) A pre-tRNAMeti molecule is depicted in the typical secondary structure shortly after transcription, with uncolored circles representing tRNA residues, pale red circles representing the 5′ leader nucleotides, pale blue circles representing the 3′ trailer nucleotides, and a bright red circle indicating the site for m1A58 modification. A pre-tRNAMeti lacking m1A58 is targeted for decay by the nucelar surveillance pathway in S. cerevisiae, involving oligoadenylation of the pre-tRNA by Trf4 of the TRAMP complex, and then 3′–5′ exonucleolytic degradation of the pre-tRNA by Rrp6 and Rrp44 of the nuclear exosome. Spliced leader-containing pre-tRNAs are also targeted for decay by the nuclear surveillance pathway (Kramer and Hopper 2013; Chatterjee et al. 2022). (Right) A mature tRNA with a CCA end is depicted in its typical secondary structure, with residues that are normally modified to form ac4C12 in yellow, m2,2G26 in blue, m7G46 in green, and m1A58 in red. Specific mature tRNAs lacking one of these modifications are targeted for decay by the rapid tRNA decay pathway in S. cerevisiae, involving 5′–3′ exonucleolytic decay of the tRNA by Rat1 and Xrn1 in the nucleus and cytoplasm, respectively, both of which are inhibited by pAp, which accumulates in met22Δ mutants.
