Figure 2.
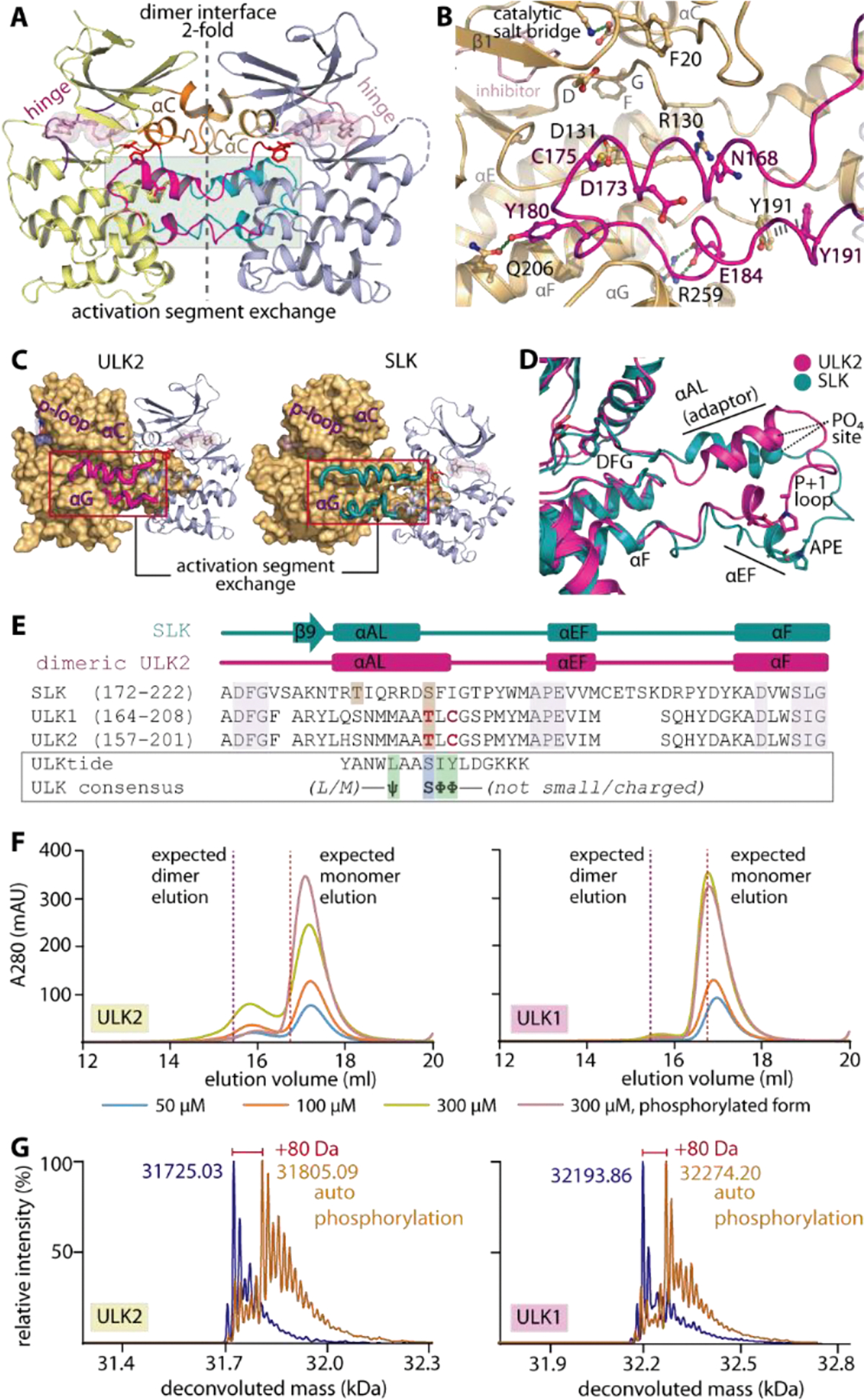
Domain-exchanged activation segment dimerization in ULK1/2. A) Face-to-face dimeric assembly in ULK2 structure showing the packing of αC and domain-exchanged activation segment at the two-fold interface. B) Detailed interactions between the protruding activation segment of one subunit and the active site of the neighboring kinase in the dimer. C) ULK2 and SLK shares similar domain-exchanged activation segment configuration for their dimerization. D) Superimposition of activation segments of ULK2 and SLK reveals a highly similar conformation. E) Structure-based sequence alignment shows that the threonine phosphorylation sites of ULK1 and ULK2 are well aligned with the S189 residue of SLK that is known to be an autophosphorylation site (brown boxes). These threonine residues are located within the region of the activation segments that lacks consensus substrate sequence, for which the residues that violate the pattern are highlighted in bold, red letters. F) Gel filtration elution profiles of ULK1 and ULK2 demonstrate potentially transient existence of dimers in solution for non-phosphorylated proteins, which decreases in the phosphorylated kinases. G) Mass spectrometry analyses reveals an increase in ~80-dalton mass upon incubating ULK1/2 with ATP and MgCl2, indicating the kinase autophosphorylation.
