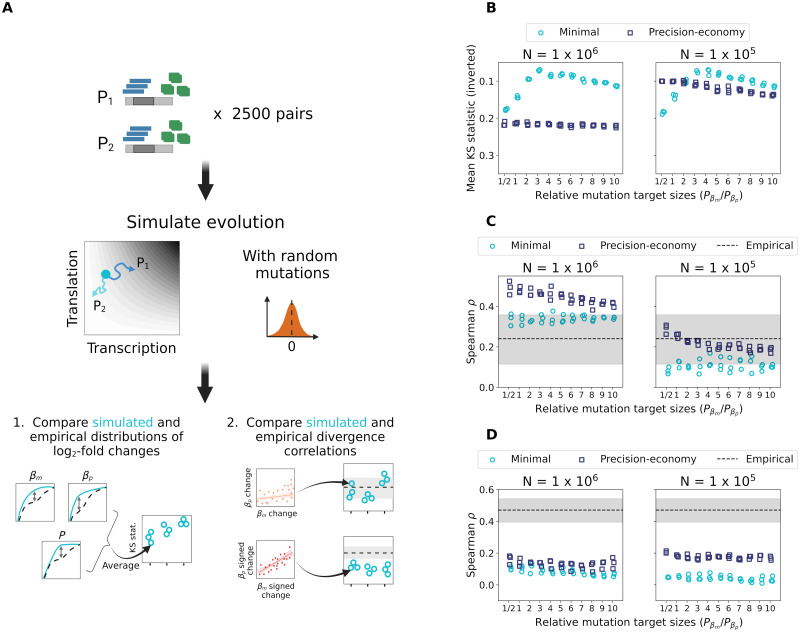
Fig 4. A larger mutational target size for transcription more robustly replicates the expression divergence patterns of yeast WGD-derived paralogs across levels of selection efficacy.
(A) For each combination of model and parameters, three replicate simulations of 2500 paralog pairs were performed. Two types of summary statistics were computed to compare simulation results to empirical observations. The Kolmogorov-Smirnov (KS) statistic, equal to the largest difference between two cumulative distribution functions, was used to quantify the distance between simulated and empirical distributions of log2-fold changes in transcription, translation and protein abundance (top). For each replicate simulation, the three resulting KS statistics were combined into a single mean value. The two divergence correlations between transcriptional and translational changes were also calculated on the set of paralog pairs obtained from each simulation, resulting in three measurements for each combination of model and parameter values (bottom). Created with BioRender.com. (B) Replication of the distributions of relative divergence in transcription rate, translation rate and protein abundance following simulations, as shown by the mean KS statistics obtained from the pairwise comparisons of all the corresponding empirical and simulated distributions. (C) Correlations between the magnitudes of transcriptional and translational log2-fold changes for gene pairs evolved in silico according to the minimal and precision-economy models. The shaded area shows the 95% confidence interval for the empirical correlation observed in S. cerevisiae WGD-derived paralogs. (D) Correlations between signed log2-fold changes in transcription and translation rates for the same simulated gene pairs. The shaded area represents the 95% confidence interval on the empirical correlation.