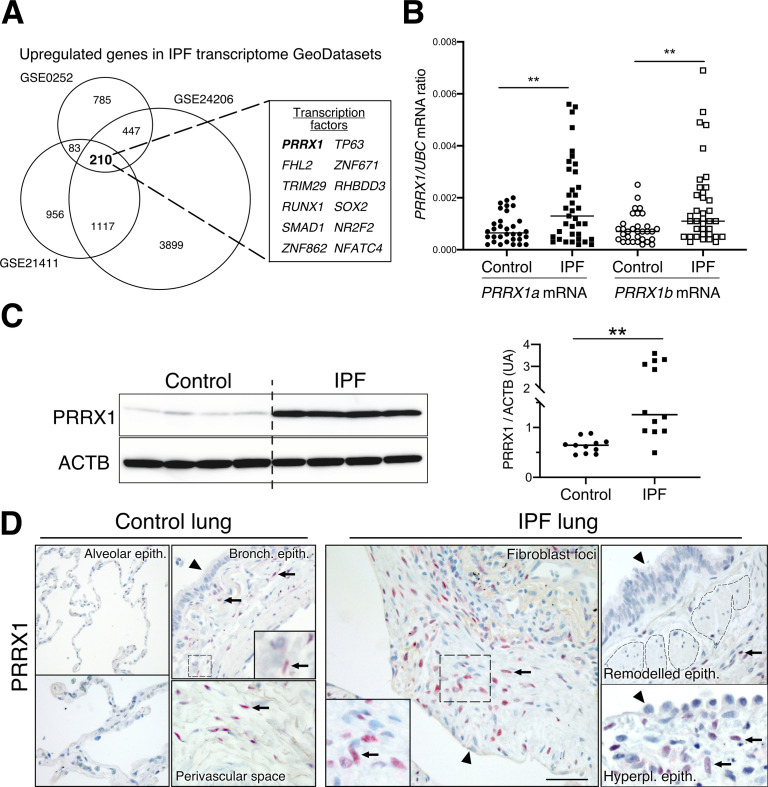
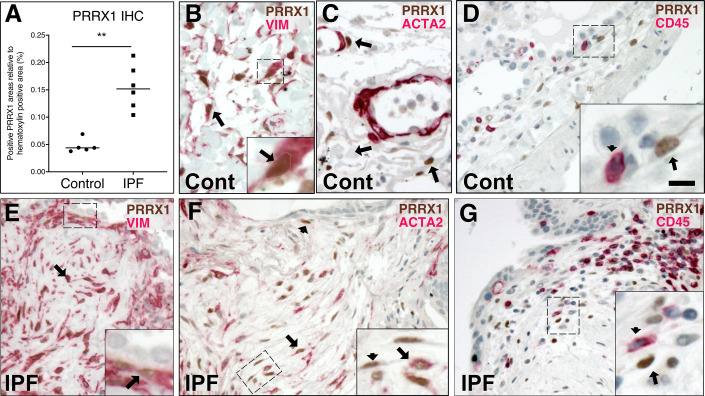
Figure 1. Identification of PRRX1 as a transcription factor reactivated in IPF lung.
(A) Venn diagram showing the number of genes up-regulated in three IPF lung Transcript microarray databases compared to controls (NCBI GEO GDS1252, GDS4279, GDS3951). Among the 210 common upregulated genes in all three datasets, 12 genes were annotated as transcription factors (see embedded table in the figure, PRRX1 is in bold). (B) Dot plots with median showing the mRNA expression of PRRX1a and PRRX1b isoforms in control (circle, n=32) and IPF (square, n=37) whole lung homogenates. (C) Immunoblot showing PRRX1 expression in control and IPF whole lung homogenates. ACTB was used as loading control. The quantification of PRRX1 relative expression to ACTB in control (circle, n=11) and IPF (square, n=12) is displayed as dot plot with median on the right. (D) Representative immunohistochemistry images (n=5 per group) showing PRRX1 staining (red) in control (left panels) and IPF (right panels). Nuclei were counterstained with hematoxylin. Note the absence of PRRX1 staining in the alveolar and bronchiolar epithelium (arrow head). PRRX1-positive cells were only detected in the peri-bronchiolar and peri-vascular spaces (arrows) in control lungs (left panels). In IPF, PRRX1-positive cells (arrow) were detected in the remodeled/fibrotic area (right panels). Note that epithelial cells (arrow head) and bronchiolar smooth muscle cells (dashed areas) are PRRX1 negative. The high-magnification pictures match the dashed boxes displayed in the main panels. Scale bar: 80 µm in low magnification images and 25 µm in high-magnification ones. Abbreviations: epithelium. (epith); bronchiolar. (bronch); Hyperpl. (hyperplastic). Mann Whitney U test, *p≤0.05, **p≤0.01.