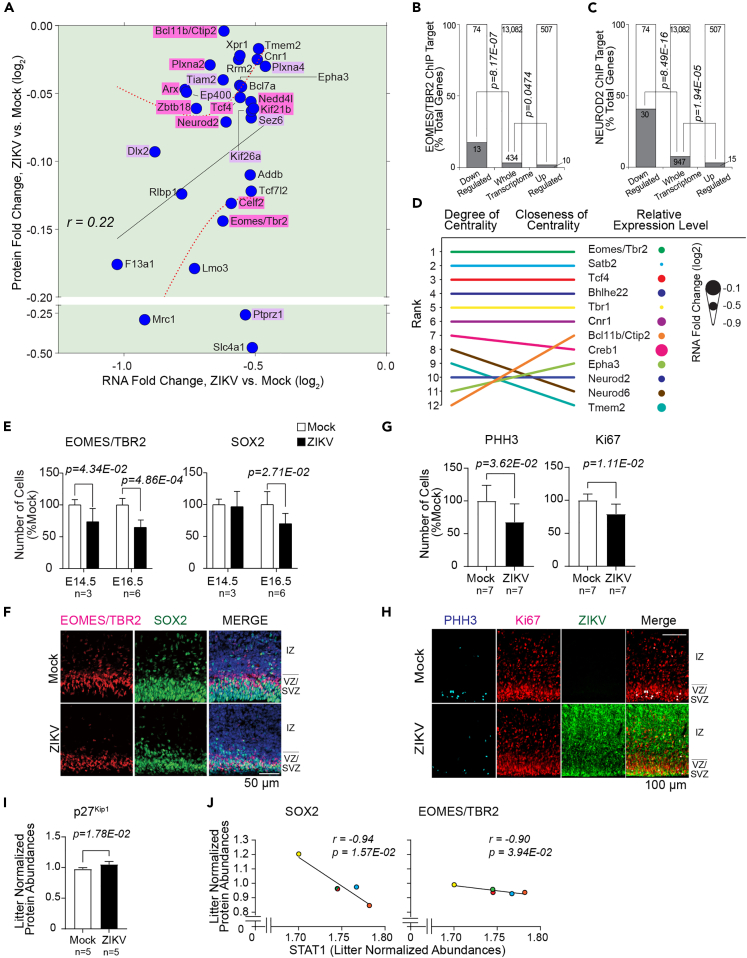
Figure 4.
ZIKV induces downregulation of critical neurodevelopmental proteins
(A) Transcript-protein pairs with decreased-in-abundance following ZIKV infection (Quadrant III of 3G). Pink; transcript-protein pairs that are known to be causative of genetic neurodevelopmental diseases in humans, Aristaless Related Homeobox (ARX), B Cell CLL/Lymphoma 11b (BCL11B or CTIP2), Eomesodermin/T-box Brain Protein 2 (EOMES/TBR2), Kinesin Family Member 21B (KIF21B), Neural Precursor Cell Expressed Developmentally Down-regulated 4-like E3 Ubiquitin Protein Ligase (NEDD4L), Neuronal Differentiation 2 (NEUROD2), Plexin A2 (PLXNA2), Transcription Factor 4 (TCF4), and Zinc Finger and BTB Domain Containing 18 (ZBTB18) (Table 2). Purple, transcript-protein pairs that are known to have neurodevelopmental functions in mice, Distal-Less Homeobox 2 (DLX2), E1A-Binding Protein P400 (EP400), Kinesin Family Member 26A (KIF26A), Plexin A4 (PLXNA4), Protein Tyrosine Phosphatase Receptor Type Z1 (PTPRZ1), Seizure Related 6 Homolog (SEZ6), and T-lymphoma invasion and metastasis-inducing protein 2 (TIAM2) (Table S7 for the functions of the highlighted transcript-protein pairs).
(B) Eomesodermin/T-box Brain Protein 2 (EOMES/TBR2) ChIP target enrichment within the down- and up-regulated subsets of the ZIKV-responsive transcriptome, relative to EOMES/TBR2 ChIP targets present in the whole transcriptome dataset; hypergeometric test p values are shown.
(C) Neuronal Differentiation 2 (NEUROD2) ChIP target enrichment within the down- and up-regulated subsets of the ZIKV-responsive transcriptome, relative to NEUROD2 ChIP targets present in the whole transcriptome dataset; hypergeometric test p values are shown.
(D) Degree and closeness of centrality and relative RNA expression level of interconnected nodes in NEUROD2/TBR2 Signaling Network. The top 10 nodes of either degree or closeness of centrality are shown.
(E) Quantification of EOMES/TBR2-positive and Sex Determining Region Y-Box 2 (SOX2)-positive cells at E14.5 and E16.5. E14.5; mock, n = 3 from 2 litters, ZIKV, n = 3 from 2 litters. E16.5; mock, n = 6 from 4 litters, ZIKV, n = 6 from 5 litters.
(F) IHC staining of VZ/SVZ and IZ of Mock- or ZIKV-infected embryonic brains, EOMES/TBR2 (red), SOX2 (green), and Hoechst (nucleus, blue), scale bar = 50 μm.
(G) Quantification of phosphoH3 (PHH3)-positive and Ki67-positive cells at E16.5; mock, n = 7 from 4 litters, zika, n = 7 from 6 litters.
(H) IHC staining of IZ and VZ/SVZ of Mock- or ZIKV-infected E16.5 brains. Mitotic marker phosphorylated Histone H3 (PHH3, cyan) and proliferation marker Ki67 (red), counterstained with ZIKV (green), Scale bar = 100 μm.
(I) Litter-normalized abundance of Cyclin-dependent Kinase Inhibitor 1B (p27Kip1) protein at E14.5.
(J) Correlation of litter-normalized abundances of neurodevelopmental genes and STAT1 abundances. Error bars indicate standard deviation.