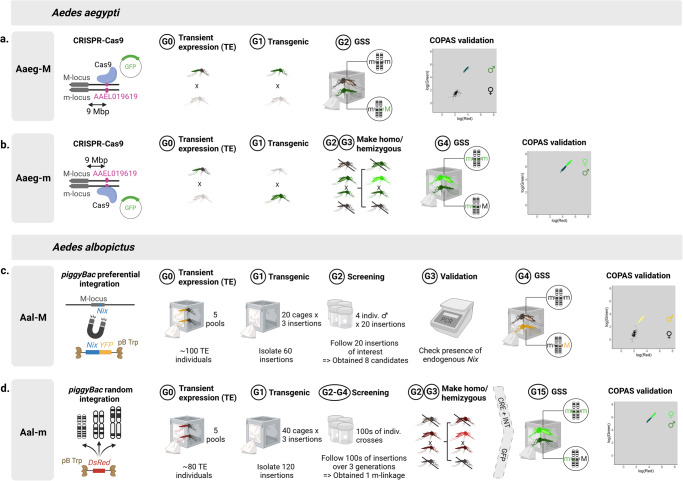
Fig. 1. Schematic of the procedures used to obtain linkage of a fluorescence marker to the M and m-loci in Ae. aegypti and Ae. albopictus.
In Ae. aegypti, a single CRISPR-Cas9 target was used to obtain an M-linked strain (a) and an m-linked strain (b) within 2–4 generations. Both lines allow sex separation using a COPAS flow cytometer, as shown on the graphs generated from representative sorting data for these lines (arbitrary fluorescence units). In Ae. albopictus, M-linkage was obtained by piggyBac preferential integration near the M-locus stimulated by the presence of Nix sequences, requiring screening and testing for 4 generations (c). “pB Trp” = helper plasmid expressing piggyBac transposase. In Ae. albopictus, m-linkage was obtained by piggyBac random integration through intensive screening over 6 generations (d). The selected m-linked line was then modified to remove undesirable transgenes using CRE-recombinase (“CRE”), an integrase (“INT”) and a GFP expressing plasmid docking into the attP site left-over after lox cassette excision. Both Aal-M and Aal-m allow sex separation using a COPAS flow cytometer (representative sorting from actual data shown, in COPAS arbitrary fluorescence units). Figure designed on BioRender.com.