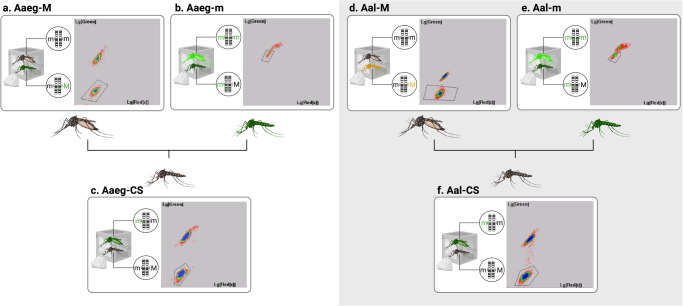
Fig. 2. Successive sortings leading to purification of non-transgenic males.
Panels show the fluorescence graphs log(Green) = f(log(Red)) displayed by the COPAS software. No scales are displayed by the software as it uses arbitrary fluorescence units. Gated regions were defined for sorting populations of interest. Of note, both GFP and YFP fluorochromes can be read in GFP mode. a Sorting of non-transgenic females from Aaeg-M strain. Total larvae = 6079. b Sorting of hemizygous transgenic males from Aaeg-m strain. Total larvae = 711. c Sorting of non-transgenic males in the progeny of 1000 non-transgenic females from Aaeg-M strain crossed with 300 hemizygous transgenic males from Aaeg-m strain. Total larvae = 65,839. d Sorting of non-transgenic females from Aal-M strain. Total larvae = 6797. e Sorting of hemizygous transgenic males from Aal-m strain. Total larvae = 1960. f Sorting of non-transgenic males in the progeny of 850 non-transgenic females from Aal-M strain crossed with 250 hemizygous transgenic males from Aal-m strain. Total larvae = 24,239. Larvae of intermediate fluorescence were observed to be either dead fluorescent larvae or larvae with lower GFP expression at the time of the sorting. All the individuals that reached the pupal stage were confirmed to be males. Figure designed on BioRender.com.