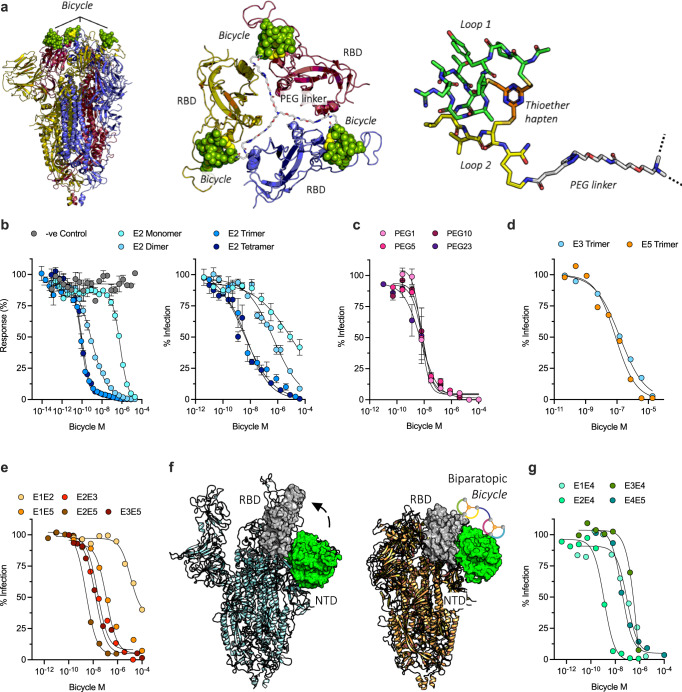
Fig. 2. Multimerisation and combinatorial assembly generates potent inhibitors.
a E2:RBD complex superposed on closed spike (PDB ID:6ZP0). Top-down view showing E2 Bicycle and RBDs from each Spike monomer (red, yellow, blue). A TCA-amido-Peg1-triazolyl linker is modeled connecting the three E2 Bicycle molecules. An individual E2 Bicycle is also shown, indicating the constraining hapten, peptide loops and PEG linker. Dotted lines indicate where other Bicycle monomers are attached via a TCA hinge. b E2 was multimerized into dimer, trimer and tetramer versions using an amido-PEG10-triazolyl linker with appropriate valency. Multimerised E2 Bicycles inhibition of Spike binding to hACE2 (left-hand graph) or Spike-pseudotyped virus (S-pV) infection of 293 T TMPRSS2/hACE2 cells (right-hand graph). Dose-response or infection curves were fit to obtain IC50 values. c E2 was trimerized with increasing TCA-amido-PEG-triazolyl linker lengths from PEG1 to PEG23. Dose-response curves for S-pV infection inhibition were fit to obtain IC50 values. d Other Spike binding, but non-ACE2 competing, Bicycles were trimerized. E3 trimer utilized a TCA-amido-PEG23-triazolyl linker and E5 trimers utilized a N-(acid-PEG3)-N-bis(PEG3-amido)-PEG36-triazolyl linker where the central hinge conjugation point was not extended by PEG36. Dose-response curves for S-pV infection inhibition were fit to obtain IC50 values: E3 = 150 nM, and E5 = 82 nM. e Dose-response curves for S-pV infection inhibition by biparatopic Bicycles utilizing a non-ACE2-competing RBD molecule. The following IC50 values were obtained: E1E2 = 11000 nM, E2E3 = 23 nM, E2E5 = 2.6 nM, E1E5 = 120 nM, and E3E5 = 16 nM. f Models of Spike in its ‘open’ (7A98) and ‘closed’ conformations (6ZP0), with the NTD and RBD domains shown as green and gray surfaces respectively. An arrow indicates that the RBD dissociates from the NTD in the open state. The expected binding of the biparatopic Bicycle E2E4 to both RBD and NTD domains is shown. g Dose-response curves for S-pV infection inhibition by biparatopic Bicycles utilizing an NTD-binding molecule. In infection experiments, the data is the result of two independent experiments presented as mean values (d, e, g) or three independent experiments as mean ± SEM (B&C). Source data are provided as a Source Data file.