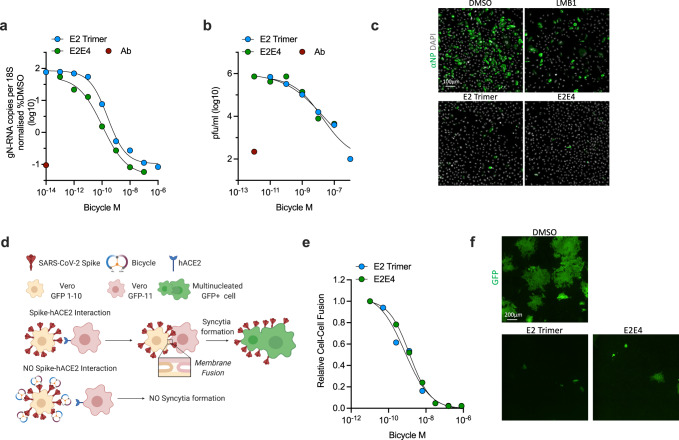
Fig. 3. Bicycles inhibit SARS-CoV-2 replication and cell–cell fusion.
a–c SARS-CoV-2 (Wuhan-Hu-1 strain) was used to infect Vero TMPRSS2/ACE2 cells in the presence of homotrimeric E2 or biparatopic E2E4. Replication was measured after 18 hours by qPCR for gN-RNA (a), 3 days by plaque assay (b) or cells were fixed and stained with anti-NP antibody (antibodies-online, ABIN6953059) after 18 hours (c). Polyclonal convalescent serum (LMB1) was used as a positive control. Data is the result of two independent experiments presented as mean values. Dose response curves were fitted to obtain qPCR IC50 values of: E2 trimer = 0.25 nM, E2E4 = 0.11 nM and plaque assay IC50 values of: E2 trimer = 1.8 nM and E2E4 = 2.0 nM. The limit of detection for plaque assays was log10(0.82) pfu/ml. d Schematic for cell–cell fusion assay. Two stable Vero cell lines each expressing one half of split GFP and either Spike or hACE2 were mixed and the formation of multinucleated syncytia monitored by the resulting GFP signal using an IncuCyte S3 live microscope. Created with Biorender.com. e Dose response curves for the inhibition of GFP+ multinucleated syncytia formation by E2 trimer and E2E4 biparatopic Bicycles, based on immunofluorescence data shown in Supplementary Figure 1B. Data is plotted as relative cell–cell fusion, where fusion in DMSO in the absence of any Bicycle, ( ~ 30% of cells), is set at 1. Fitting the data results in IC50 values of: E2 trimer = 0.74 nM and E2E4 = 1.3 nM. Data shown is from two independent experiments presented as mean values. f Immunofluorescence images of GFP+ multinucleated syncytia formed in the presence of DMSO, 32 nM E2 trimer or 32 nM E2E4 Bicycles. Data is representative of two independent experiments. Source data are provided as a Source Data file.