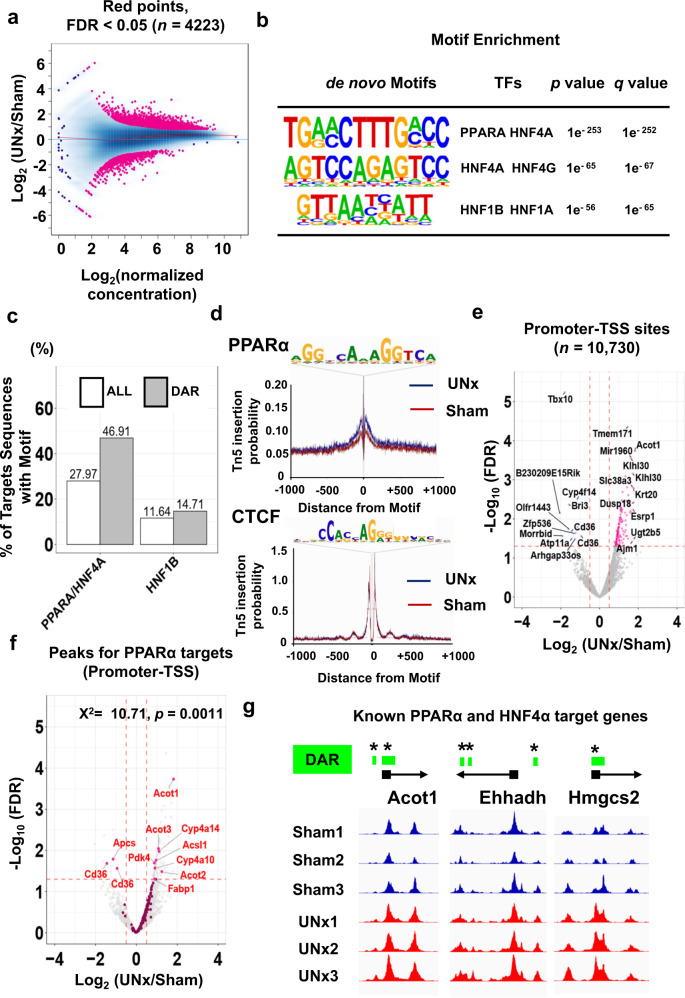
Fig. 3. Single tubule ATAC-seq for microdissected proximal tubules from Sham and UNx at the 24 h timepoint.
a Ratio-intensity plot. Points represent peak regions determined by MACS2 for individual peaks. The x-axis represents average signal intensity within the region, and the y-axis represents log2 ratio of Sham and UNx signals. Red line is Loess fit. Among the 125,973 open chromatin regions identified among all samples, 4223 were identified as differentially accessible sites (FDR < 0.05, highlighted in magenta). (n = 3 for each group) b HOMER analysis identifies the enriched TF binding motifs in chromatin regions that are more accessible in UNx vs Sham (applying cumulative hypergeometric distribution adjusted for multiple testing with the Benjamini-Hochberg method) (See also Supplementary Data 5 for all results). c Percentages of motif sequences in all peaks (white) or in differentially accessible regions (grey) for the PPARα/HNF4α target motif and the HNF1B target motif. d Transcription factor foot-printing profiles generated using all identified ATAC-seq peaks for PPARα and CTCF (negative control) for microdissected PT-S1 to quantitate Tn5 insertion enrichment in the UNx vs. sham. e Volcano plot indicating peaks of chromatin accessibility within annotated promoter-TSS regions. Magenta points, increased accessibility in the UNx vs Sham treatment (FDR < 0.05 and log2(UNx/Sham)> 0.5). Blue points, decreased accessibility (FDR < 0.05 and log2(UNx/Sham) < −0.5). Chromatin accessible regions with top 10 or bottom 10 log2(UNx/Sham) values annotated by nearest gene name. f Volcano plot indicating chromatin accessibility at promoter regions near annotated TSS. Magenta points, PPARα target regions. Target genes for PPARα are listed in Supplementary Data 7. g Examples of ATAC-seq peaks for known PPARα and HNF4α target genes, each showing differences in peak heights between UNx and Sham treatments at their promoter-TSS regions (highlighted in green; DAR, differentially accessible region, vertical axes are of equal length). All data are available on a genome browser at https://esbl.nhlbi.nih.gov/IGV_mo/ and a Shiny-based web page (https://esbl.nhlbi.nih.gov/UNx/). * FDR < 0.05 (Benjamini and Hochberg method, adjusted p values are provided in Supplementary Data 6).