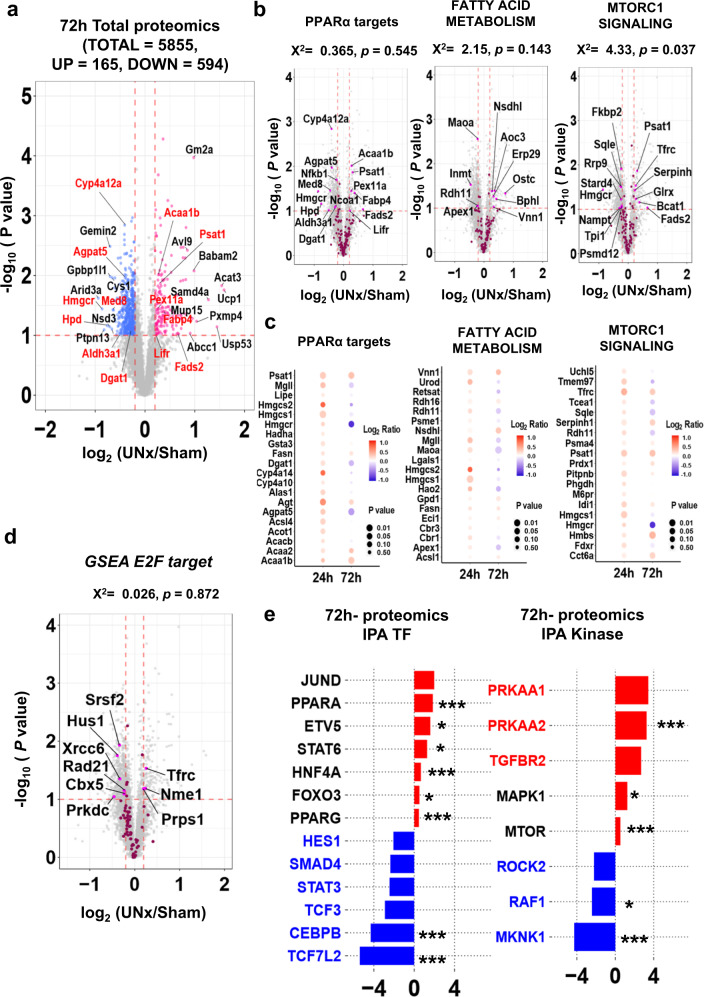
Fig. 8. Quantitative proteomics of whole kidneys from Sham and UNx at the 72 h timepoint.
a Volcano plot for UNx vs. Sham. TMT-based quantitative proteomics LC-MS/MS of whole kidneys from mice with either Sham (n = 4) or UNx (n = 4) surgery. Red dots, upregulated proteins upregulated in UNx (p < 0.1 and log2 (UNx/Sham) > 0.2); blue dots, downregulated proteins in UNx (p < 0.1 and log2 (UNx/Sham) < −0.2). PPARα regulated proteins in red font. b Volcano plots highlighting proteins known to be regulated by PPARα, and proteins with roles in “FATTY ACID METABOLISM” and “MTORC1_SIGNALING” in GSEA indicated by magenta points. p-values associated with enrichment of respective gene sets (Chi-squared analysis). c Bubble plots showing differential changes of proteins between the 24 h and 72 h time points for the proteins regulated by PPARα, and for the proteins annotated as “FATTY ACID METABOLISM” and “MTORC1_SIGNALING” in GSEA. P values (unpaired, two-tailed t-test) are visualized by circle size, and log2 ratios are visualized by color. d Volcano plot for the proteins annotated as “E2F TARGET” in GSEA. Magenta points, known E2F targets. Significant differential abundance was determined using thresholds of p < 0.1 (unpaired, two-tailed T-test) and|log2 (UNx/Sham)| > 0.2. p-value represents the likelihood of E2F target protein enrichment among regulated peaks using Chi-squared analysis. e Prediction of upstream regulatory transcription factors (left) and kinases (right) determined using Ingenuity Pathway Analysis (IPA) at 72 h. *p < 0.05, **p < 0.01, ***p < 0.001 (Fisher’s Exact Test, p values are provided in Supplementary Data 21).