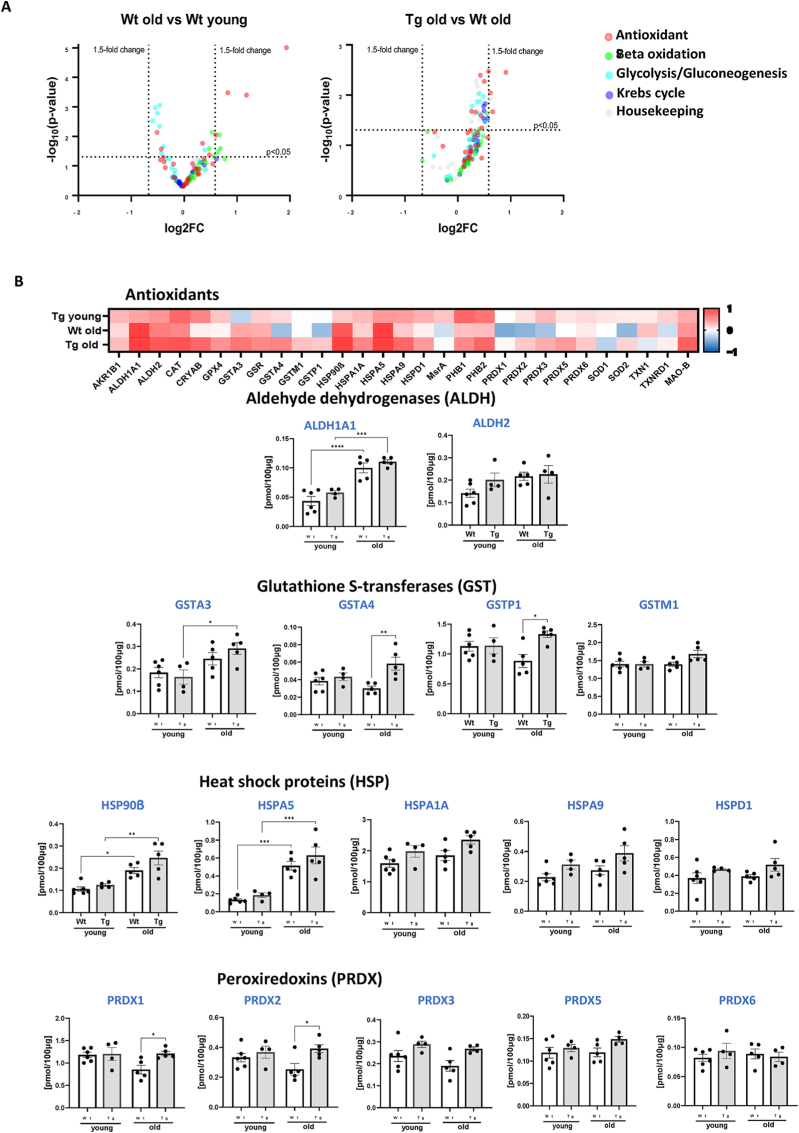
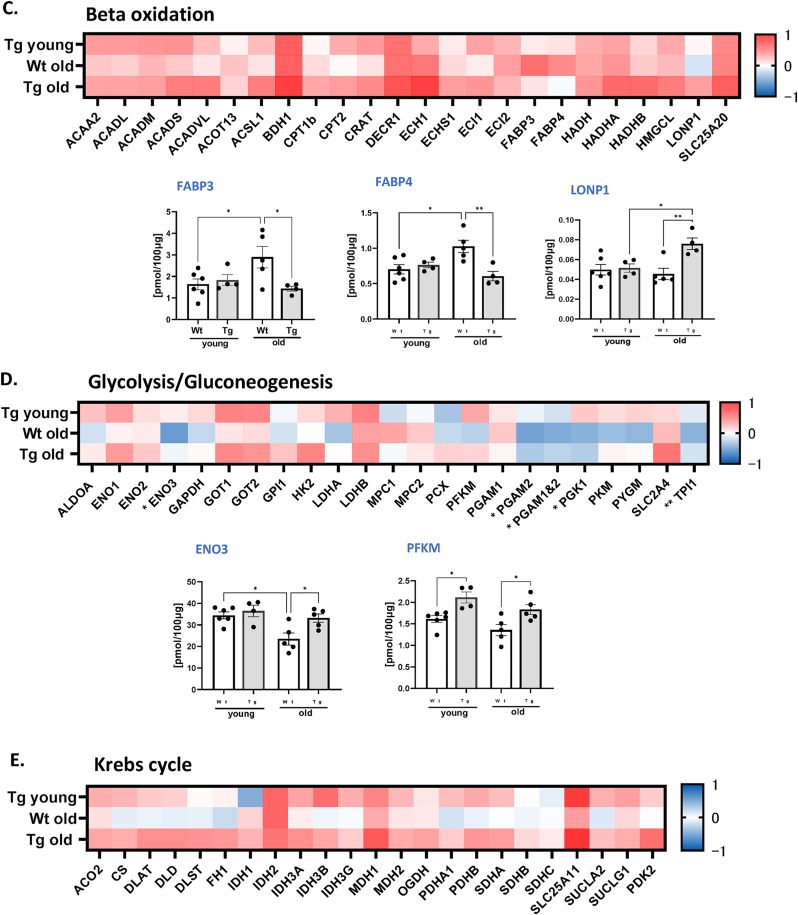
Fig. 9.
Targeted proteomic analysis of key metabolic and antioxidant proteins. (A) Volcano plot of the proteins showing the Log2 fold changes plotted against corresponding –log10 p-value for wildtype old mice versus young, and GPX4Tg versus wildtype old. Data points in the upper right (ratio >1.5) and upper left (ratio <0.67, 1.5- fold decrease) sections with P > 0.05 represent proteins that are significantly changed. (B) Heatmap representing the log2 fold change in the levels of antioxidant proteins and theconcentration of selected proteins from the antioxidant panel: Aldehyde dehydrogenases (ALDH): ALDH1A1 and ALDH2; Glutathione S-transferases (GST): GSTA3, GSTA4, GSTP1, and GSTM1; Heat shock proteins (HSP): HSP90β, HSPA5, HSPA1A, HSP9, HSPD1; Peroxiredoxins (PRDX): PRDX1, PRDX2, PRDX3, PRDX4, PRDX5. (C) Heatmap representing the log2 fold change values of proteins level and the concentration of selected proteins from the (C) beta oxidation panel: fatty acid binding protein 3 (FABP3), fatty acid binding protein 4 (FABP4) and lon peptidase 1 (LONP1); (D) glycolysis/gluconeogenesis panel: enolase 3 (ENO3) and phosphofructokinase muscle-type (PFKM) and (D) heatmap for krebs cycle panel. *Significant difference between the groups (p < 0.05, one-way ANOVA), n = 4–6.