Figure 1.
The strategy of the quantitative phagosomal lumen acidification assay
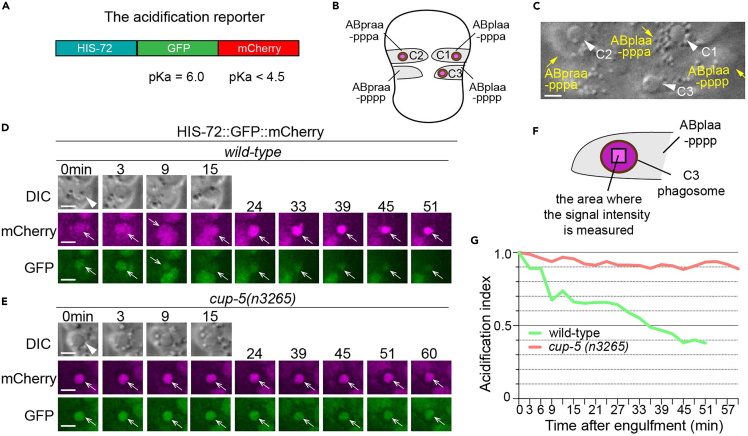
(A) Domain structure of the HIS-72::GFP::mCherry reporter construct. The pKa value of each individual fluorescent protein is indicated.
(B) A diagram illustrating the three phagosomes that contain cell corpses C1, C2, and C3, with which we monitor the acidification process, at ∼330 min post the 1st embryonic division. Both the positions of C1, C2, and C3 (brown dots) and the identities of their engulfing cells are shown.
(C) An image showing how C1, C2, and C3 (arrowheads) can be distinguished under DIC objectives. The scale bar is 5 μm.
(D and E) Time-lapse imaging series of phagosomes (white arrowheads in DIC images) of wild-type and cup-5 mutant embryos expressing Phis-72his-72::gfp::mCherry. Open white arrows depict the nuclei of engulfed cell corpses, labeled with both the GFP and mCherry markers. Reduction of the GFP signal intensity over time is indicative of phagosome acidification. ‘0 min’ is when a phagosome is just sealed. Scale bars are 2 μm.
(F) A diagram illustrating where the intensities of the mCherry and GFP signals are measured in the center of a phagosome.
(G) The acidification index curves of two phagosomes (Y-axis) over time (in the 3 min interval) (X-axis) in embryos with the labeled genotypes. ‘0 min’ indicates the moment when a phagosome is just sealed. The data of the wild-type and cup-5 (n3265) are from D-E, respectively.
(B, F, D, E, G) are adapted from Figure 1 (B and C) and Figure 13 (A, B, and D) of Pena-Ramos et al.1