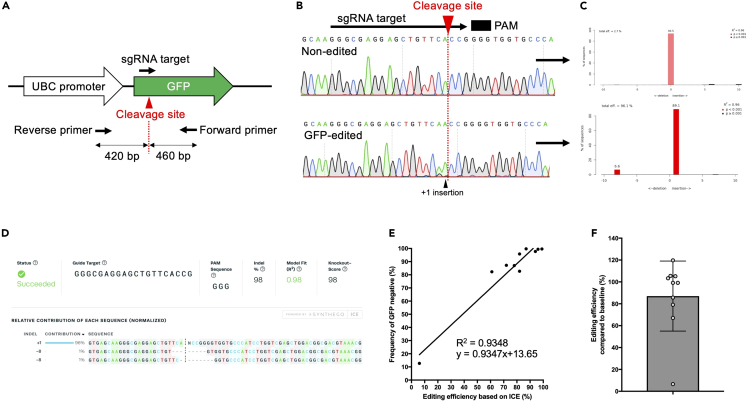
Figure 3.
Primer design for evaluating genome editing efficiency
(A) Design of sequence primer sets for sgRNA targeting of the UBC-GFP locus.
(B) Representative Sanger sequencing results of non-edited and GFP-edited cells.
(C) Results of a TIDE assay.
(D) Results of ICE analysis.
(E) Genome editing efficiency based on ICE positively correlated with GFP expression levels. The donor-derived peripheral blood cells from individual recipient mice in two independent transplant experiments were used for ICE analysis (mean ± SD, n = 10).
(F) The ratio of genome editing efficiency at the transplant to genome editing efficiency at four months after transplant. The same data set is used as in Figure 2E (mean ± SD, n = 10).