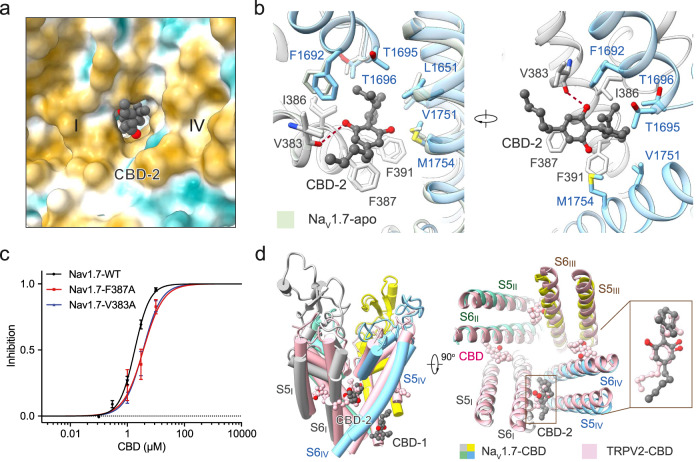
Fig. 4. Coordination of CBD at the F-site.
a CBD binds to the fenestration enclosed by repeats IV and I of Nav1.7. The surrounding environment, which is highly hydrophobic, is shown as the hydrophobic surface, calculated in ChimeraX. b The local structures around the F-site (left) remain nearly identical in apo (PDB: 7W9K) and CBD-bound Nav1.7. Surrounding residues are shown as sticks. The only potential hydrogen bond is shown as red dashed lines. c F-site mutations weaken the inhibition of Nav1.7 by CBD. Both single point mutations V383A and F387A reduced the sensitivity of Nav1.7 to CBD, with the IC50 right-shifted from 1.82 ± 0.10 μM to 3.56 ± 0.58 μM and 3.65 ± 0.78 μM, respectively. Nav1.7-WT, n = 1, 5, 12, 8, 7. Nav1.7-F387A, n = 4, 6, 4. Nav1.7–V383A, n = 5, 4,5. Data are presented as mean ± SEM. n biological independent cells. d CBD shares similar binding poses in Nav1.7 and TRPV2 at the F-site. A similar fenestration binding site for CBD in Nav1.7 and in TRPV2 is seen in the superimposed structures. A side view (left) and a top view (right) of the comparison between CBD-bound Nav1.7 (domain colored) and TRPV2 (pink, PDB: 6U88) are shown.