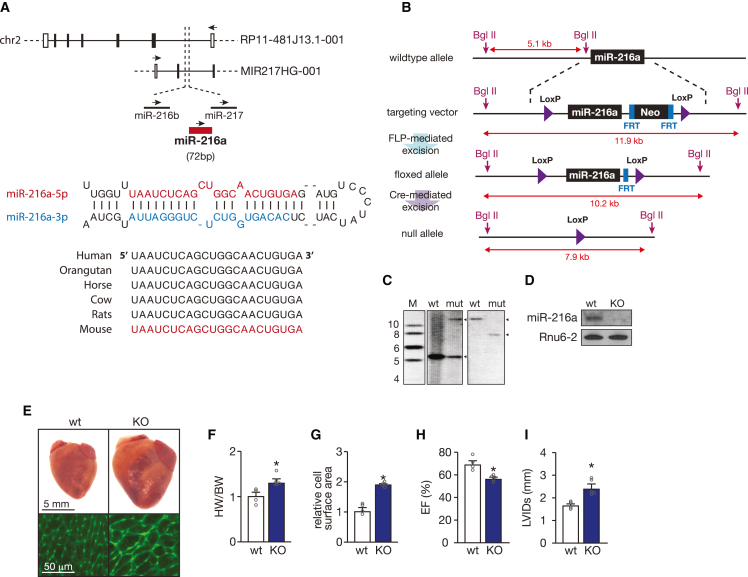
Figure 2.
miRNA-216a genomic location and targeting strategy
(A) Schematic representation of hsa-miR-216a-5p genomic localization (top panel) and precursor sequence (bottom panel). In the human genome, miR-216a is in a cluster together with miR-216b and miR-217, which is transcribed together within a long non-coding RNA MIR217HG-001 within the opposite strand of a longer long non-coding RNA RP11-481J13.1-001. The mature miR-216a-5p strand is conserved among species (bottom panel). (B) Strategy for targeting of miR-216a. The targeting vector, targeted allele, and null allele for miR-216a are shown. LoxP sites were introduced flanking the genomic region encompassing both the 71-bp pre-miR-216a as well as a neomycin resistance cassette flanked by two FRT sites, which allowed for bFLP recombinase-mediated excision and Cre-mediated removal of the targeted region of the miR-216a locus. Sizes of probes for Southern blotting analysis are shown. (C) Verification of homologous recombination strategy by Southern blot analysis based on BgII digestion. The wild-type (WT) allele yielded a 5.1 kb DNA fragment, whereas successful targeting (mut) results in both 5.1 and 11.9 kb fragments in heterozygous miR-216aneo/+. Removal of the neomycin cassette and Cre-mediated excision of the floxed miR-216a target region, an expected fragment of 7.9 kb was observed (null allele). (D) Northern blotting analysis of miR-216a-5p expression in hearts from WT and miR-216a knockout (KO) mice. Rnu6-2 was used as a loading control. (E) Representative images of whole hearts (top panel) and wheat germ agglutinin (WGA)-stained (bottom panel) histological sections of WT and KO hearts. Scale bars, 5 mm (top panel) and 50 μm (bottom panel). (F) Gravimetric analysis of corrected heart weights in WT and KO mice; n refers to number of hearts: n = 4 (WT) and n = 5 (KO). (G) Quantification of cardiomyocyte surface area from WT and KO mice, based on 150 cells measured per heart; n refers to number of hearts: n = 4 (WT) and n = 5 (KO). (H) Quantification of ejection fraction (EF) and (I) left ventricular internal diameter (LVID) during systole in WT and KO mice; n refers to number of hearts: n = 4 (WT) and n = 5 (KO). In all panels, numerical data are presented as mean (error bars show SEM); statistical significance was calculated using two-tailed unpaired t test. ∗p < 0.05 vs. control group.