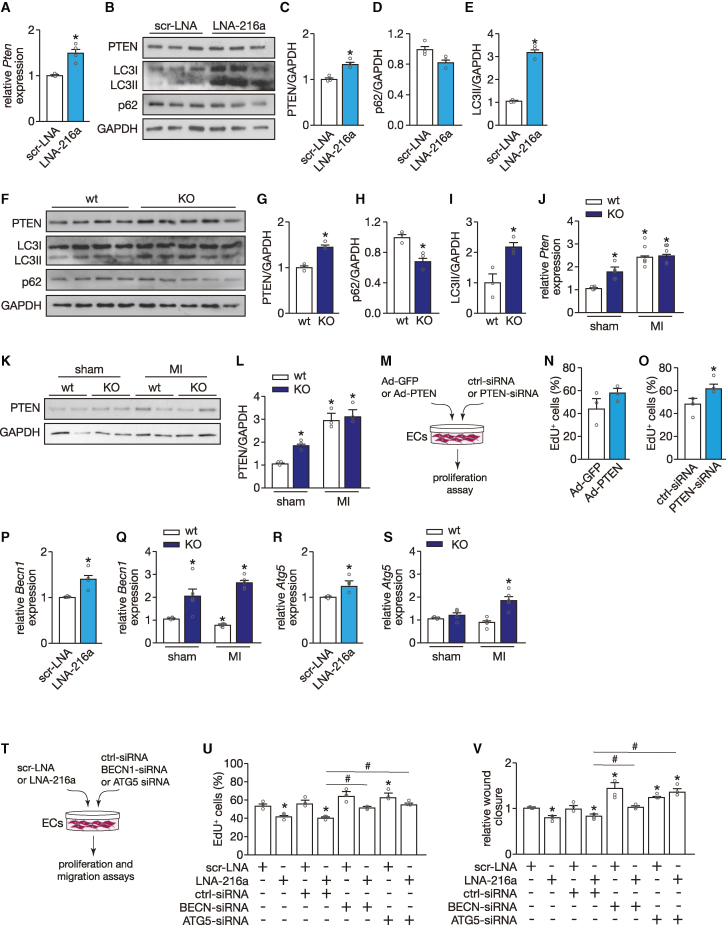
Figure 6.
miR-216a links cardiac endothelial function and autophagy via regulation of PTEN
(A) Quantitative PCR analysis of PTEN in HUVECs transfected either with a scrambled LNA or an LNA-miR-216a (n = 4 independent experiments including each condition). (B) Western blot analysis of PTEN and two autophagy surrogate markers, LC3 and p62, in HUVECs transfected either with a scrambled LNA or an LNA-miR-216a. GAPDH was used as loading control. (C–E) Quantification of PTEN expression (C), p62 (D), and LC3-II (E) from conditions in (B); n = 3 independent experiments including each condition. (F) Western blot analysis of PTEN, LC3, and p62 in myocardial tissue of WT and KO mice. GAPDH was used as loading control. (G–I) Quantification of PTEN expression (G), p62 (H), and LC3-II (I) in experimental groups from (G); n = 3 independent experiments including each condition. (J) Quantitative PCR analysis of Pten in myocardial tissue of WT and KO mice subjected to sham or MI; n = 5 hearts per group. (K) Western blot analysis and (L) quantification of PTEN expression in myocardial tissue of WT and KO mice subjected to sham or MI; n = 3 hearts per group. (M) Experimental setup of modulation of PTEN expression levels in HUVECs, using a specific siRNA to achieve downregulation and an adenovirus for upregulation. (N) Quantification of HUVEC proliferation 48 h after transduction with an Ad-GFP (control) or an Ad-PTEN, represented as percentage of Edu-positive cells; n = 3 independent experiments (10 microscopic fields for each condition/experiment). (O) Quantification of HUVEC proliferation 48 h after transfection with a scrambled- or PTEN-siRNA, represented as percentage of Edu-positive cells; n = 3 independent experiments (10 microscopic fields for each condition/experiment). (P) Quantitative PCR analysis of BECN1 in HUVECs transfected either with a scrambled LNA or an LNA-miR-216a (n = 4 independent experiments including each condition). (Q) Quantitative PCR analysis of Becn1 in myocardial tissue of WT and KO mice subjected to sham or MI; n = 5 hearts per group. (R) Quantitative PCR analysis of ATG5 in HUVECs transfected either with a scrambled LNA or an LNA-miR-216a (n = 4 independent experiments including each condition). (S) Quantitative PCR analysis of Atg5 in myocardial tissue of WT and KO mice subjected to sham or MI; n = 5 hearts per group. (T) Experimental setup to inhibit either BECN1 or ATG5 expression levels in HUVECs upon downregulation of miR-216a, using a specific siRNA and an LNA probe, respectively. (U) Quantification of HUVEC proliferation 48 h after transfection with scrambled LNA, or an LNA-miR-216, and ctrl-sirna, BECN1-sirna or ATG5-siRNA, represented as percentage of Edu-positive cells; n = 3 independent experiments (10 microscopic fields for each condition/experiment). (V) Quantification of wound healing/migration assay on HUVECs after transfection with scrambled LNA, or an LNA-miR-216, and ctrl-sirna, BECN1-sirna or ATG5-siRNA, represented as wound closure rate (n = 3 independent experiments for each condition). In all panels numerical data are presented as mean (error bars show SEM); statistical significance was calculated using two-tailed unpaired t test when comparing two experimental groups or two-way ANOVA followed by Tukey’s multiple comparison test when comparing more than two experimental groups. ∗p < 0.05 vs. corresponding control group.