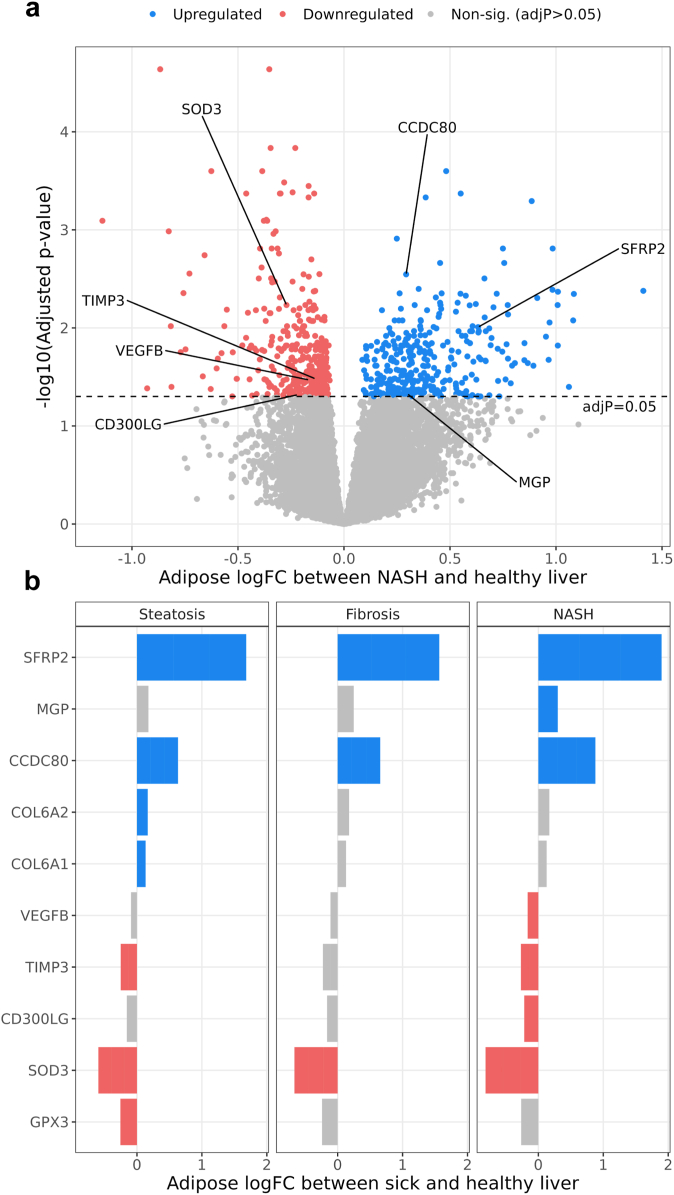
Fig. 2.
A total of 953 genes are differentially expressed (DE) in subcutaneous adipose tissue between the obese individuals with the three main non-alcoholic fatty liver disease (NAFLD) traits, steatosis, fibrosis and/or non-alcoholic steatohepatitis (NASH), and the obese individuals with healthy livers. We performed DE analysis on bulk RNA-seq data from subcutaneous adipose biopsies in the KOBS cohort, comparing individuals with the NAFLD traits diagnosed by liver histology to those with healthy livers. Gene counts represent numbers of genes DE for NAFLD in the subcutaneous adipose tissue before filtering for serum biomarker candidates (SBCs). Of the 953 adipose DE genes, 680, 273, and 663 genes are DE for steatosis, fibrosis, and NASH, respectively. (a) Volcano plot showing the results of the NASH DE analysis in the adipose tissue. The X-axis represents log fold-change (logFC) in adipose bulk RNA-seq data from individuals with NASH and those with healthy livers. The Y-axis represents the negative log of the DE p-value, adjusted for multiple testing with the Benjamini-Hochberg procedure. Significant SBCs identified in our subsequent filtering steps (Fig. 3) are highlighted. Volcano plots of steatosis and fibrosis DE results in the subcutaneous adipose tissue are shown in Supplementary Fig. S3. (b) Bar plot showing the DE direction of the SBCs in the adipose DE analysis for steatosis, fibrosis, and NASH. X-axis represents logFC in adipose bulk RNA-seq data from individuals with each NAFLD trait and those with healthy livers. Y-axis represents the SBC name, sorted by logFC. Blue SBCs have increased adipose expression in individuals with NAFLD when compared to the individuals with healthy livers, while red SBCs have decreased adipose expression.