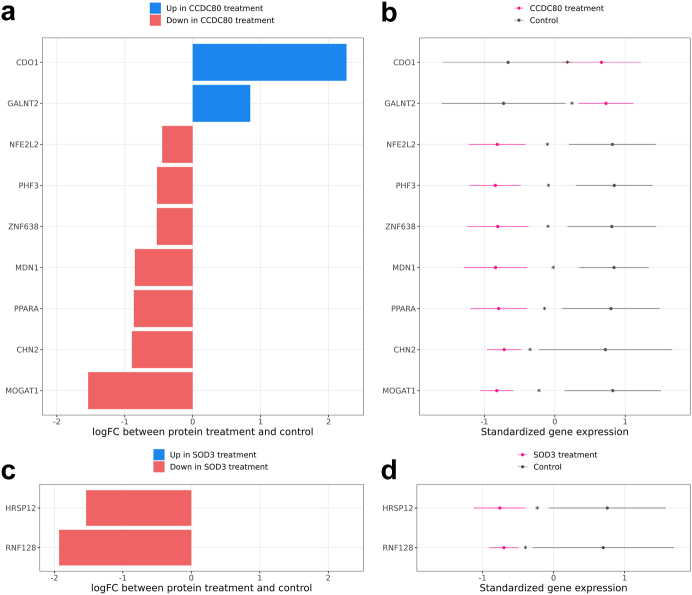
Fig. 6.
Treatment of human liver HepG2 cells with the CCDC80 and SOD3 recombinant proteins changes the expression of several known NAFLD-related genes. We treated separate cultures of human liver HepG2 cells with the CCDC80 and SOD3 recombinant proteins for 24 h (see Methods), and measured expression via RNA-seq. We then performed a differential expression (DE) analysis between the recombinant protein treated cells and non-treated control cells. (a) Results of the CCDC80 treatment DE analysis. The X-axis represents the log fold-change (logFC) of the 9 significant DE genes, and the Y-axis represents the gene names. Blue genes were expressed significantly more in the CCDC80 treated cells than in the control cells, and red genes were expressed less. (b) Mean expression of all 9 significant DE genes in the CCDC80 treated cells compared to the control cells. The X-axis represents counts per million (CPM), standardized by the mean and standard deviation of each gene; the Y-axis gene name; error bars the mean ± one standard deviation; and colours the experimental condition (CCDC80 treated HepG2 cells or non-treated control cells). Each row represents 4 samples treated with CCDC80 and 4 control samples, for a total of 8 samples per row. The “∗” annotation indicates that the gene was significantly DE between the CCDC80 treatment and control samples (adjP < 0.05 after Benjamini–Hochberg correction (FDR < 0.05)). (c) Results of the SOD3 treatment DE analysis, with 2 significant DE genes. Plot elements are analogous to those in (a). (d) Mean expression of the 2 DE genes in the SOD3 treated cells compared to the control cells. Plot elements are analogous to those in (b).