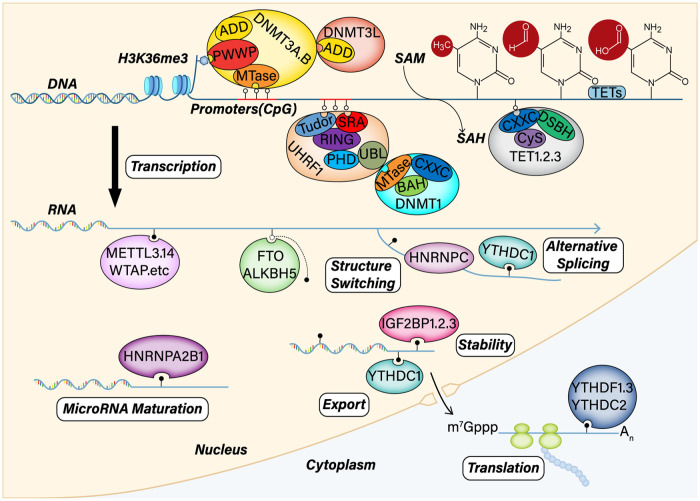
Figure 1.
DNA and RNA methylation machinery and mechanisms. DNA methylation modification includes three phases: de novo DNA methylation, maintenance methylation, and demethylation. DNMT3A and DNMT3B are responsible for de novo DNA methylation, which contain a highly conserved DNMT domain (the MTase domain) in the carboxy terminus and two chromatin reading domains, ADD and PWWP. DNMT3L also comprises ADD and interacts with and stimulates the activity of DNMT3A and DNMT3B. DNMT1 is critical for maintaining DNA methylation after replication through cell division. UHRF1 is also responsible for maintaining methylation. UHRF1 specifically binds CpG dinucleotides at replication forks through its SRA domain and H3K9me2 and H3K9me3 through its Tudor-PHD domain. UHRF1 recruits DNMT1 through its UBL domain, thereby relieving its auto-inhibition. The TET family carries out DNA demethylation via the oxidation of methylcytosine. The TET family includes TET1/2/3, which contain catalytic domains (Cysh-rich and DSBH). However, TET1 and TET3 also have CXXC regions. As a post transcriptional modification, m6A can influence RNA processing and metabolism via various methods, including alternative splicing, structure switching, stability, maturation, translocation, and translation.