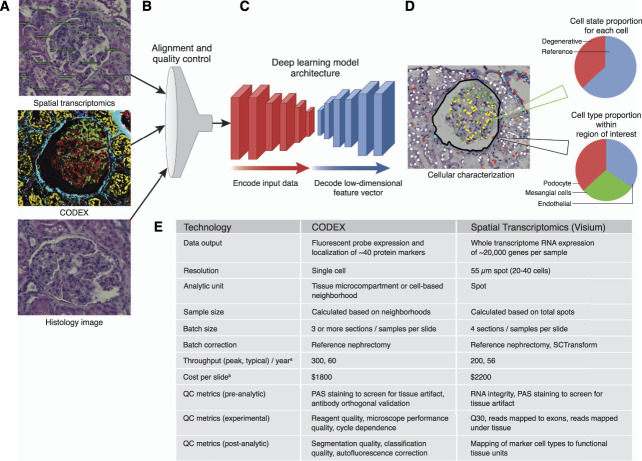
Figure 1.
Deep learning for fusion of two popular spatial technologies. A deep learning model using diverse spatial -omics data as the input and spatial mapping of -omics data on bright-field histology as the output. (A) Glomerulus image from varying modalities including brightfield histology, overlaid spot locations for Visium Spatial Transcriptomics, and Co-Detection by Indexing (CODEX) image. (B) The alignment and quality control step ensures accurate registration of spatial-omics data as well as read quality of transcriptomic or fluorescence data. (C) Representative deep learning model architecture consisting of two sets of convolutional filter banks to first compress input data into a low-dimensional vector and then decode that low-dimensional input and render predictions. (D) Cellular characterization as the output of an ML model, providing insights into both cell-type composition for a given region of interest and an estimate of cellular health (cell state). (E) Comparison of spatial technologies. ML, machine learning; PAS, periodic acid–Schiff; QC, quality control.