Figure 4. Homology modeling of key mutations in XBB.1.5, CH.1.1, and CA.3.1.
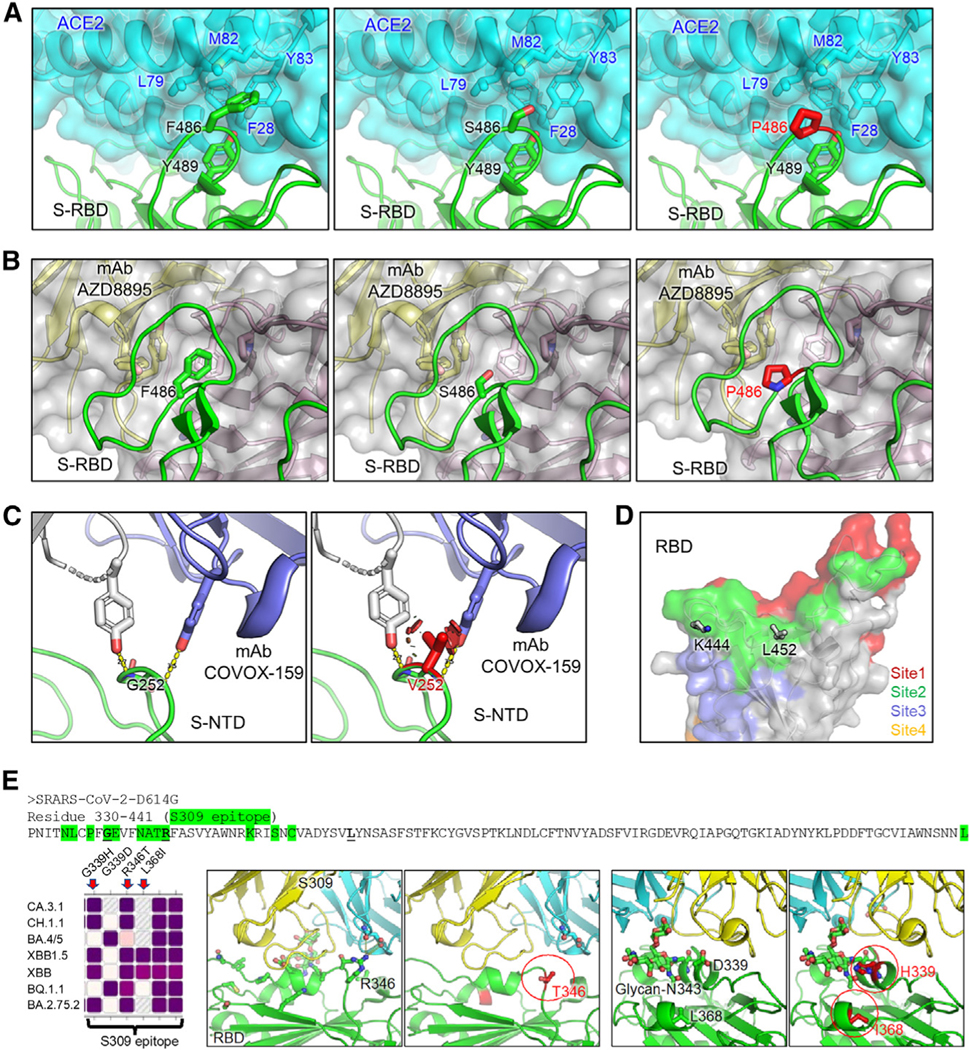
(A) Structures of S receptor-binding domain (RBD)-ACE2 binding interface shown as ribbons.
(B) Structure of RBD with class I antibody AZD8895. The recognition focuses on residue F486, with multiple antibody residues forming a surrounding hydrophobic cage, whereas this interaction is abolished by F486S/P mutation.
(C) Structures of an immune-dominant region of S N-terminal domain (NTD) with a representative antibody COVOX-159. The nAb recognition on residue G252 is abolished by G252V mutation through creating a steric hindrance (shown as red plates).
(D) Residues K444 and L452 are located within acommon epitope site of class II RBD-targeting neutralizing antibodies represented as green surface.
(E) Antibody S309 epitope and sequence diversity.(Top) S protein sequence (330–441) with residues of antibody S309 epitope highlighted in green, and mutation hotspots in bold font, (left) amino acid variation at residues 339, 346, and 368 among different Omicron subvariants, (middle) R346T abolishes a salt bridge and a hydrogen bond; and (right) G/D339H interferes with the S309 recognition of glycan-N343 while L368I stabilizes it.
