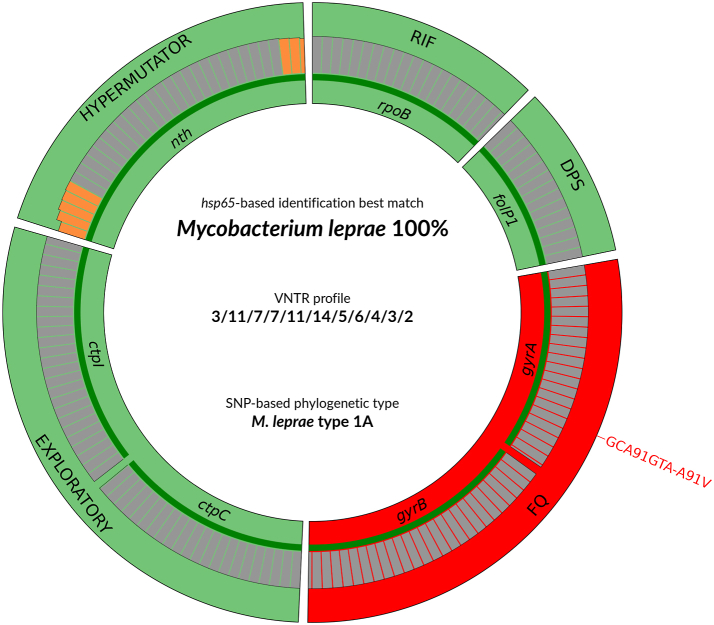
Fig. 1.
Deeplex Myc-Lep results identifying a M. leprae strain of SNP type 1A genotypically resistant to fluoroquinolones. Results are shown for a Thai53 strain derivative, mutated in gyrA (see Table S3). Information on hsp65 best match-based identification, VNTR allelic profile and SNP-based phylogenetic type is shown in the center of the circle. Information on predictions of drug susceptibility and drug resistance for anti-leprosy drugs/drug classes and on hypermutator genotype is as follows. Target gene regions are grouped within sectors in a circular map according to the prediction feature (drug resistance, hypermutation) with which they are associated. Sectors in red and green indicate targets in which resistance- or hypermutation-associated mutations or no mutations are detected, resulting in predictions of resistant or susceptible phenotypes (for rpoB, folP1, gyrA, gyrB), or hypermutator strain (nth), respectively. The ctpC and ctpI sector (and their associated drug resistance or drug susceptibility predictions) are categorized as exploratory, based on previous work suggesting an association of missense mutations in these genes with resistance to rifampicin, observed in a single strain devoid of mutation in the rpoB DRDR (see text). Green lines above gene names represent the reference sequences with coverage breadth above 95%. Limit of detection (LOD) of minority variants (resulting from subpopulations of reads bearing a mutation) depends on the read depth at each sequence position and is shown either as grey (LOD 10%) or orange zones (LOD >10%) above reference sequences. Here, LOD is >10% at the extremities of the nth target only. In the VNTR profile, VNTR markers are ordered as follows: 6-3a, AC8a, AC8b, AC9, GTA9, GAA21, GGT5, 6–7, 12-5, 21-3, 23-3. ∗RIF: Rifampicin, DPS: Dapsone, FQ: Fluoroquinolones, VNTR, variable-number tandem-repeat, SNP, single nucleotide polymorphism.