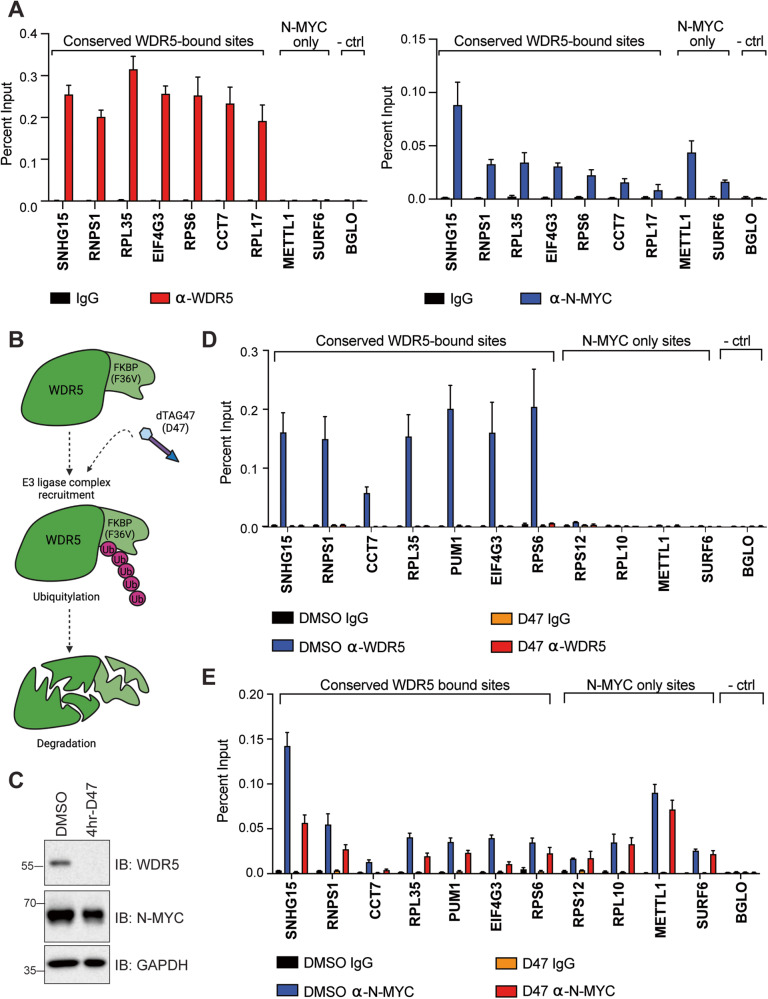
Fig. 2. Impact of WDR5 depletion on the ability of N-MYC to bind chromatin in N-MYC amplified neuroblastoma cells.
A Co-immunoprecipitated DNA from ChIP for N-MYC and WDR5 in IMR-32 cells was probed by QPCR using primers targeting 7 loci that are conserved WDR5-bound sites and 2 that are not classified as WDR5-bound. BGLO is a negative control locus. Data represented as a percentage of input DNA for each locus (n = 3 biological replicates, error bars are standard error). B CHP-134 cells were engineered previously to express a version of WDR5 fused to the FKBP(F36V) degron [15] (“called DTWDR5 cells”). In these cells, the addition of dTAG47 (D47) links WDR5 to an E3 ligase complex, resulting in ubiquitylation and subsequent rapid degradation through the proteosome system. Image created with BioRender.com. C Western blot showing the level of WDR5 expression in DTWDR5 cells following 4 h treatment with 500 nM D47 compared to matched DMSO control. GAPDH is used to control loading. D DTWDR5 cells were treated as in (C) and ChIP was performed for WDR5 following extraction of chromatin. Graph shows that chromatin-bound WDR5 is absent upon D47 treatment at 7 loci that are conserved WDR5-bound sites. E DTWDR5 cells were treated as in (C) and ChIP was performed for N-MYC. At sites where WDR5 is absent (D), N-MYC binding is reduced. For both (D) and (E), BGLO is a negative control locus. Data represented as a percentage of input DNA for each locus (n = 3 biological replicates, error bars are standard error).