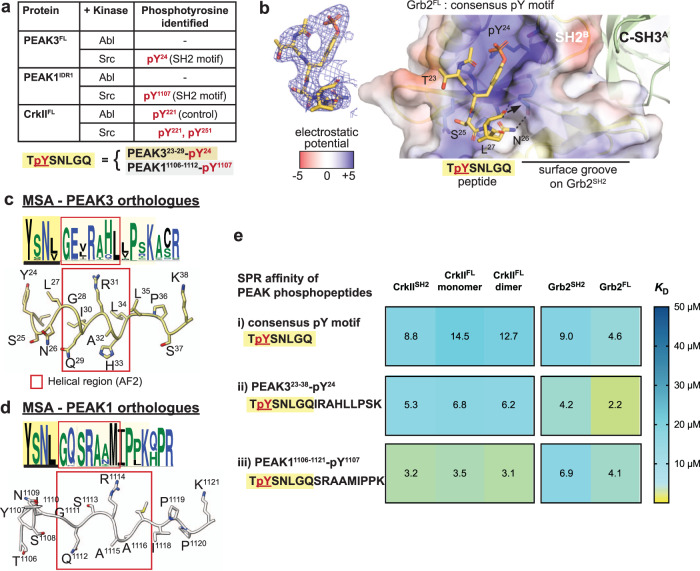
Fig. 2. Structural and biophysical analysis of PEAK/Grb2SH2 interaction.
a TYSNL site of PEAK3FL (Y24) and PEAK1IDR1 (Y1107) can be phosphorylated by Src (MS/MS analysis of tryptic peptides following in vitro kinase assay of PEAK3FL and PEAK1IDR1 are available in source data). Both Abl and Src can phosphorylate CrkIIY221, a known Abl phosphorylation site84, demonstrating that both Abl and Src kinases are active. b Zoom in structure of the PEAK SH2-pY peptide bound to the Grb2FL dimer, with the PEAK phosphopeptide shown in yellow and Grb2FL Chain B (peptide bound chain) shown in surface representation, colored by electrostatic surface potential calculated using the APBS Electrostatics plugin85 for PyMOL (Schroedinger) (blue = positive, white = hydrophobic, red = negative). Inset shows a 2Fo-Fc electron density map (blue mesh, contoured at 1.0σ) showing the final modeled PEAK3/PEAK1 consensus phosphopeptide (TpYSNLGQ, yellow sticks, underlined residues modeled). See also Supplementary Fig. 2. c, d PEAK323–38 and PEAK11106–1121 regions alongside full MSA/Web Logo of orthologs for the corresponding sequence, showing a section of high sequence conservation (pale yellow box) adjacent to the TYSNL SH2 site (yellow box, underlined) including a region predicted in AF2 to adopt helical secondary structure (red box). e Tabulated SPR-determined mean steady-state binding affinity values (KD) for PEAK phosphotyrosine peptides (consensus pY-SH2 motif and extended peptides PEAK323-38pY24 and PEAK11106–1121-pY1107) binding to CrkIISH2, full-length CrkII (CrkIIFL) monomer or dimer, Grb2SH2 or full length Grb2 (Grb2FL). Values are colored as a heat map to illustrate trends in affinity. Data represent mean values from n = 3 independent titrations. See Supplementary Fig. 2d for representative SPR sensorgrams and Supplementary Data 1 for full SPR tabulated data and sensorgrams.