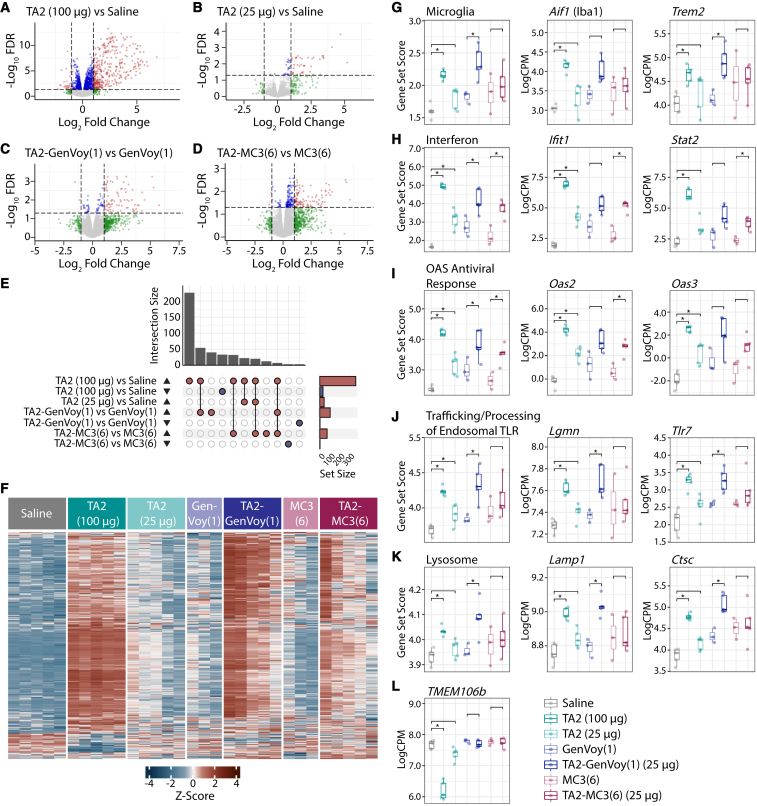
Figure 5.
Transcriptomic analysis characterizing ASO-LNP-mediated gliosis
(A–D) Volcano plots of differential gene expression profiles from pairwise comparisons of TA2 (100 μg) vs. saline (A), TA2 (25 μg) vs. saline (B), TA2-GenVoy(1) vs. GenVoy(1) (C), and TA2-MC3(6) vs. MC3(6) (D). Red dots represent FDR < 0.05 and absolute log fold change (logFC) > 1, blue dots represent FDR < 0.05 and absolute logFC < 1, green dots represent FDR > 0.05 and absolute logFC > 1, and gray dots represent FDR > 0.05 and absolute logFC < 1. (E) UpSet plot of direction stratified overlaps between significant (FDR < 0.05 and absolute logFC > 1) differentially expressed genes (DEGs) from all pairwise comparisons. (F) Heatmap of hierarchical clustering analysis of significant (FDR < 0.05 and absolute logFC > 1) DEGs from all pairwise comparisons. (G–K) Gene set scores and select genes for microglia (G), interferon (H), OAS antiviral response (I), trafficking and processing of endosomal TLR (J), and lysosome (K). (L) TMEM106b mRNA expression levels. Gene set score comparisons were analyzed using unpaired two-tailed Student’s t test, ∗p < 0.05. Individual gene comparisons were calculated using voom-limma, ∗FDR < 0.05.